3-benzyl-1-(trans-4-((5-cyanopyridin-2-yl)amino)cy. clohexyl)-1-arylurea derivatives as novel and. selective cyclin-dependent kinase 12 (CDK12)
|
|
|
- Frigyes Nagy
- 6 évvel ezelőtt
- Látták:
Átírás
1 SUPPORTING INFORMATION Discovery of 3-benzyl-1-(trans-4-((5-cyanopyridin-2-yl)amino)cy clohexyl)-1-arylurea derivatives as novel and selective cyclin-dependent kinase 12 (CDK12) inhibitors Masahiro Ito,*, Toshio Tanaka,, # Akinori Toita, Noriko Uchiyama, Hironori Kokubo,, # Nao Morishita, Michael G. Klein, Hua Zou, Morio Murakami, Mitsuyo Kondo,,# Tomoya Sameshima, Shinsuke Araki, Satoshi Endo,, Tomohiro Kawamoto,,# Gregg B. Morin,, Samuel A. Aparicio, Atsushi Nakanishi, Hironobu Maezaki,,# and Yasuhiro Imaeda*, Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited, 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa , Japan Department of Structural Biology, Takeda California Inc., Science Center Dr., San Diego, CA 92121, United States Genome Sciences Centre, British Columbia Cancer Agency, 675 W. 10th Avenue, Vancouver, BC, V5Z 1L3, Canada S1
2 Department of Medical Genetics, University of British Columbia, Vancouver, BC, V6H 3N1, Canada Department of Molecular Oncology, British Columbia Cancer Agency, 675 W. 10th Avenue, Vancouver, BC, V5Z 1L3, Canada Corresponding Author * (M.I.) phone: , masahiro.ito@takeda.com; (Y.I.) phone: , yasuhiro.imaeda@takeda.com Present Addresses # For T. Tanaka, H. Kokubo, M. Kondo, T. Kawamoto, and H. Maezaki: Axcelead Drug Discovery Partners Inc., Fujisawa-shi, Kanagawa, , Japan. For S. Endo: Academic Research and Industrial Collaboration Management Office, Kyushu University, Momochihama, Sawara-ku, Fukuoka, , Japan. TABLE OF CONTENTS Supporting information 1 (Overlay of co-crystal structures (PDB:4NST and PDB: 6CKX)).. S3 Supporting information 2 (The global kinase selectivity of 2)... S4 9 Supporting information 3 (Time dependency for CDK12 inhibition)... S10 11 Supporting information 4 (Co-crystal structure of 29 and CDK12)... S12 16 Supporting information 5 (Single crystal X-ray structure analyses)... S17 22 References... S23 S2
3 Supporting Information 1 (Overlay of co-crystal structures (PDB:4NST and PDB: 6CKX)) Supplementary Figure 1. Overlay of co-crystal structures (PDB:4NST, ligand: ADP, orange; PDB: 6CKX, ligand: compound 29, cyan). The C-terminal extension of CDK12 (PDB:4NST) associates with the bound nucleotide in the ATP cleft. In contrast, the C-terminal extension in the complex of CDK12 and compound 29 was disordered and not defined (PDB:6CKX) and the phenylpyrazole moiety in compound 29 extends out toward the solvent region in which the C-terminal extension could reside. S3
4 Supporting Information 2 (The global kinase selectivity of 2) Z-lyte Life Technologies' SelectScreen Profiling Service: Single Point Results SelectScreen Scientist: Derek Johnston Date: SSBK-Z'-LYTE (Paisley UK) Quality Assurance Review: Andrew Heuze Date: Legend < 40% Inhibition % Phosphorylation Pass 40% - 80% Inhibition Z' Determination Pass 80% Inhibition Project # Compound No. Development 1X Test Compound Difference Between Reaction Concentration [ATP] Tested Kinase Tested % Inhibition % Inhibition Data Points Interference Test Compound Interference (nm) (μm) Point 1 Point 2 mean Point 1 - Point 2 Coumarin Fluorescein SSBK9686_ Km app ABL1 E255K Pass Pass Pass 0.89 PV3864/34528 SSBK9686_ Km app ABL1 G250E Pass Pass Pass 0.90 PV3865/34529 SSBK9686_ Km app ABL1 T315I Pass Pass Pass 0.84 PV3866/39639 SSBK9686_ Km app ABL1 Y253F Pass Pass Pass 0.88 PV3863/34531 SSBK9686_ Km app ABL Pass Pass Pass 0.77 P3049/ SSBK9686_ Km app ABL2 (Arg) Pass Pass Pass 0.86 PV3266/ SSBK9686_ Km app ACVR1B (ALK4) Pass Pass Pass 0.88 PV4312/ SSBK9686_ Km app ADRBK1 (GRK2) Pass Pass Pass 0.85 PV3361/ SSBK9686_ Km app ADRBK2 (GRK3) Pass Pass Pass 0.91 PV3827/38897 SSBK9686_ Km app AKT1 (PKB alpha) Pass Pass Pass 0.85 P2999/ SSBK9686_ Km app AKT2 (PKB beta) Pass Pass Pass 0.87 PV3184/28770 SSBK9686_ Km app AKT3 (PKB gamma) Pass Pass Pass 0.88 PV3185/28771 SSBK9686_ Km app ALK Pass Pass Pass 0.86 PV3867/ SSBK9686_ Km app AMPK A1/B1/G Pass Pass Pass 0.84 PV4672/ SSBK9686_ Km app AMPK A2/B1/G Pass Pass Pass 0.70 PV4674/ SSBK9686_ Km app AURKA (Aurora A) Pass Pass Pass 0.68 PV3612/32155 SSBK9686_ Km app AURKB (Aurora B) Pass Pass Pass 0.90 PV6130/ SSBK9686_ Km app AURKC (Aurora C) Pass Pass Pass 0.74 PV3856/ SSBK9686_ Km app AXL Pass Pass Pass 0.77 PV3971/ SSBK9686_ Km app BLK Pass Pass Pass 0.74 PV3683/33635 SSBK9686_ Km app BMX Pass Pass Pass 0.78 PV3371/ SSBK9686_ BRAF V599E Pass Pass Pass 0.63 PV3849/ SSBK9686_ BRAF Pass Pass Pass 0.81 PV3848/ SSBK9686_ Km app BRSK1 (SAD1) Pass Pass Pass 0.89 PV4333/36097 SSBK9686_ Km app BTK Pass Pass Pass 0.81 PV3363/ SSBK9686_ Km app CAMK1D (CaMKI delta) Pass Pass Pass 0.82 PV3663/ SSBK9686_ Km app CAMK2A (CaMKII alpha) Pass Pass Pass 0.80 PV3142/28192 SSBK9686_ Km app CAMK2B (CaMKII beta) Pass Pass Pass 0.81 PV4205/35330 SSBK9686_ Km app CAMK2D (CaMKII delta) Pass Pass Pass 0.78 PV3373/31647 SSBK9686_ Km app CAMK4 (CaMKIV) Pass Pass Pass 0.68 PV3310/ SSBK9686_ Km app CDC42 BPA (MRCKA) Pass Pass Pass 0.67 PV4398/ SSBK9686_ Km app CDC42 BPB (MRCKB) Pass Pass Pass 0.78 PV4399/36845 SSBK9686_ Km app CDK1/cyclin B Pass Pass Pass 0.91 PV3292/ SSBK9686_ Km app CDK2/cyclin A Pass Pass Pass 0.82 PV3267/ SSBK9686_ Km app CDK5/p Pass Pass Pass 0.91 PV4676/ SSBK9686_ Km app CDK5/p Pass Pass Pass 0.83 PV3000/25348 SSBK9686_ Km app CHEK1 (CHK1) Pass Pass Pass 0.70 P3040/28702 SSBK9686_ Km app CHEK2 (CHK2) Pass Pass Pass 0.74 PV3367/ SSBK9686_ Km app CLK Pass Pass Pass 0.83 PV3315/ SSBK9686_ Km app CLK Pass Pass Pass 0.86 PV4201/ SSBK9686_ Km app CLK Pass Pass Pass 0.92 PV3826/ SSBK9686_ Km app CSF1R (FMS) Pass Pass Pass 0.80 PV3249/ SSBK9686_ Km app CSK Pass Pass Pass 0.90 P2927/ SSBK9686_ Km app CSNK1A1 (CK1 alpha 1) Pass Pass Pass 0.91 PV3850/ SSBK9686_ Km app CSNK1D (CK1 delta) Pass Pass Pass 0.85 PV3665/ SSBK9686_ Km app CSNK1E (CK1 epsilon) Pass Pass Pass 0.85 PV3500/ SSBK9686_ Km app CSNK1G1 (CK1 gamma 1) Pass Pass Pass 0.72 PV3825/34360 SSBK9686_ Km app CSNK1G2 (CK1 gamma 2) Pass Pass Pass 0.83 PV3499/ SSBK9686_ Km app CSNK1G3 (CK1 gamma 3) Pass Pass Pass 0.80 PV3838/ SSBK9686_ Km app CSNK2A1 (CK2 alpha 1) Pass Pass Pass 0.81 PV3248/ SSBK9686_ Km app CSNK2A2 (CK2 alpha 2) Pass Pass Pass 0.76 PV3624/32653 SSBK9686_ Km app DAPK3 (ZIPK) Pass Pass Pass 0.79 PV3686/ SSBK9686_ Km app DCAMKL2 (DCK2) Pass Pass Pass 0.82 PV4297/ SSBK9686_ Km app DNA-PK Pass Pass Pass 0.79 PV5864/ SSBK9686_ Km app DYRK1A Pass Pass Pass 0.89 PV3785/ SSBK9686_ Km app DYRK1B Pass Pass Pass 0.82 PV4649/ SSBK9686_ Km app DYRK Pass Pass Pass 0.88 PV3837/ SSBK9686_ Km app DYRK Pass Pass Pass 0.88 PV3871/37361 SSBK9686_ Km app EEF2K Pass Pass Pass 0.84 PV4559/ SSBK9686_ Km app EGFR (ErbB1) L858R Pass Pass Pass 0.87 PV4128/ SSBK9686_ Km app EGFR (ErbB1) L861Q Pass Pass Pass 0.68 PV3873/34562 SSBK9686_ Km app EGFR (ErbB1) T790M L858R Pass Pass Pass 0.90 PV4879/ SSBK9686_ Km app EGFR (ErbB1) T790M Pass Pass Pass 0.52 PV4803/ SSBK9686_ Km app EGFR (ErbB1) Pass Pass Pass 0.79 PV3872/ SSBK9686_ Km app EPHA Pass Pass Pass 0.76 PV3841/ SSBK9686_ Km app EPHA Pass Pass Pass 0.90 PV3688/ SSBK9686_ Km app EPHA Pass Pass Pass 0.86 PV3651/32933 SSBK9686_ Km app EPHA Pass Pass Pass 0.84 PV3840/34383 SSBK9686_ Km app EPHA Pass Pass Pass 0.92 PV3844/36870 SSBK9686_ Km app EPHB Pass Pass Pass 0.88 PV3786/34225 SSBK9686_ Km app EPHB Pass Pass Pass 0.86 PV3625/ SSBK9686_ Km app EPHB Pass Pass Pass 0.91 PV3658/33066 SSBK9686_ Km app EPHB Pass Pass Pass 0.90 PV3251/29241 SSBK9686_ Km app ERBB2 (HER2) Pass Pass Pass 0.91 PV3366/ SSBK9686_ Km app ERBB4 (HER4) Pass Pass Pass 0.88 PV3626/32657 SSBK9686_ Km app FER Pass Pass Pass 0.91 PV3806/38946 SSBK9686_ Km app FES (FPS) Pass Pass Pass 0.82 PV3354/35734 SSBK9686_ Km app FGFR Pass Pass Pass 0.79 PV3146/ SSBK9686_ Km app FGFR Pass Pass Pass 0.90 PV3368/31517 SSBK9686_ Km app FGFR3 K650E Pass Pass Pass 0.86 PV4392/36445 SSBK9686_ Km app FGFR Pass Pass Pass 0.87 PV3145/28459 SSBK9686_ Km app FGFR Pass Pass Pass 0.79 P3054/26967 SSBK9686_ Km app FGR Pass Pass Pass 0.79 P3041/ SSBK9686_ Km app FLT1 (VEGFR1) Pass Pass Pass 0.89 PV3666/ SSBK9686_ Km app FLT3 D835Y Pass Pass Pass 0.76 PV3967/ SSBK9686_ Km app FLT Pass Pass Pass 0.78 PV3182/ SSBK9686_ Km app FLT4 (VEGFR3) Pass Pass Pass 0.87 PV4129/38454 SSBK9686_ Km app FRAP1 (mtor) Pass Pass Pass 0.70 PV4753/ SSBK9686_ Km app FRK (PTK5) Pass Pass Pass 0.71 PV3874/ SSBK9686_ Km app FYN Pass Pass Pass 0.86 P3042/ SSBK9686_ Km app GRK Pass Pass Pass 0.86 PV3807/ Z' Kinase Part# / Lot# S4
5 SSBK9686_ Km app GRK Pass Pass Pass 0.88 PV3824/ SSBK9686_ Km app GRK Pass Pass Pass 0.85 PV3661/37437 SSBK9686_ Km app GRK Pass Pass Pass 0.93 PV3823/34013 SSBK9686_ Km app GSK3A (GSK3 alpha) Pass Pass Pass 0.86 PV6126/ SSBK9686_ Km app GSK3B (GSK3 beta) Pass Pass Pass 0.79 PV3365/ SSBK9686_ Km app HCK Pass Pass Pass 0.85 PV6128/ SSBK9686_ Km app HIPK1 (Myak) Pass Pass Pass 0.89 PV4561/ SSBK9686_ Km app HIPK Pass Pass Pass 0.92 PV5275/ SSBK9686_ Km app HIPK3 (YAK1) Pass Pass Pass 0.89 PV4209/ SSBK9686_ Km app HIPK Pass Pass Pass 0.90 PV3852/ SSBK9686_ Km app IGF1R Pass Pass Pass 0.83 PV3250/35957 SSBK9686_ Km app IKBKB (IKK beta) Pass Pass Pass 0.60 PV3836/ SSBK9686_ Km app IKBKE (IKK epsilon) Pass Pass Pass 0.63 PV4875/ SSBK9686_ Km app INSR Pass Pass Pass 0.90 PV3781/ SSBK9686_ Km app INSRR (IRR) Pass Pass Pass 0.64 PV3808/34272 SSBK9686_ Km app IRAK Pass Pass Pass 0.57 PV3362/37360 SSBK9686_ Km app ITK Pass Pass Pass 0.70 PV3875/ SSBK9686_ Km app JAK Pass Pass Pass 0.80 PV4774/ SSBK9686_ Km app JAK2 JH1 JH2 V617F Pass Pass Pass 0.82 PV4336/ SSBK9686_ Km app JAK2 JH1 JH Pass Pass Pass 0.72 PV4393/ SSBK9686_ Km app JAK Pass Pass Pass 0.83 PV4210/36301 SSBK9686_ Km app JAK Pass Pass Pass 0.78 PV3855/ SSBK9686_ Km app KDR (VEGFR2) Pass Pass Pass 0.81 PV3660/ SSBK9686_ Km app KIT T670I Pass Pass Pass 0.58 PV3869/34504 SSBK9686_ Km app KIT Pass Pass Pass 0.71 P3081/ SSBK9686_ Km app LCK Pass Pass Pass 0.81 P3043/ SSBK9686_ Km app LTK (TYK1) Pass Pass Pass 0.74 PV4651/ SSBK9686_ Km app LYN A Pass Pass Pass 0.84 P2906/ SSBK9686_ Km app LYN B Pass Pass Pass 0.79 P2907/21076 SSBK9686_ MAP2K1 (MEK1) Pass Pass Pass 0.75 PV3303/ SSBK9686_ MAP2K2 (MEK2) Pass Pass Pass 0.81 PV3615/32519 SSBK9686_ MAP2K6 (MKK6) Pass Pass Pass 0.88 PV3318/ SSBK9686_ MAP3K8 (COT) Pass Pass Pass 0.76 PV4313/ SSBK9686_ Km app MAP3K9 (MLK1) Pass Pass Pass 0.76 PV3787/ SSBK9686_ Km app MAP4K2 (GCK) Pass Pass Pass 0.74 PV4211/ SSBK9686_ Km app MAP4K4 (HGK) Pass Pass Pass 0.66 PV3687/ SSBK9686_ Km app MAP4K5 (KHS1) Pass Pass Pass 0.64 PV3682/ SSBK9686_ Km app MAPK1 (ERK2) Pass Pass Pass 0.89 PV3313/ SSBK9686_ MAPK10 (JNK3) Pass Pass Pass 0.76 PV4563/ SSBK9686_ Km app MAPK11 (p38 beta) Pass Pass Pass 0.82 PV3679/ SSBK9686_ Km app MAPK12 (p38 gamma) Pass Pass Pass 0.82 PV3654/ SSBK9686_ Km app MAPK13 (p38 delta) Pass Pass Pass 0.86 PV3656/ SSBK9686_ Km app MAPK14 (p38 alpha) Direct Pass Pass Pass 0.87 PV3304/ SSBK9686_ MAPK14 (p38 alpha) Pass Pass Pass 0.75 PV3304/ SSBK9686_ Km app MAPK3 (ERK1) Pass Pass Pass 0.77 PV3311/ SSBK9686_ MAPK8 (JNK1) Pass Pass Pass 0.78 PV3319/ SSBK9686_ MAPK9 (JNK2) Pass Pass Pass 0.53 PV3620/32388 SSBK9686_ Km app MAPKAPK Pass Pass Pass 0.86 PV3317/ SSBK9686_ Km app MAPKAPK Pass Pass Pass 0.83 PV3299/38895 SSBK9686_ Km app MAPKAPK5 (PRAK) Pass Pass Pass 0.80 PV3301/ SSBK9686_ Km app MARK1 (MARK) Pass Pass Pass 0.81 PV4395/ SSBK9686_ Km app MARK Pass Pass Pass 0.89 PV3878/ SSBK9686_ Km app MARK Pass Pass Pass 0.78 PV4819/ SSBK9686_ Km app MARK Pass Pass Pass 0.63 PV3851/ SSBK9686_ Km app MATK (HYL) Pass Pass Pass 0.89 PV3370/31553 SSBK9686_ Km app MELK Pass Pass Pass 0.88 PV4823/ SSBK9686_ Km app MERTK (cmer) Pass Pass Pass 0.86 PV3627/32658 SSBK9686_ Km app MET (cmet) Pass Pass Pass 0.67 PV3143/ SSBK9686_ Km app MET M1250T Pass Pass Pass 0.51 PV3968/34718 SSBK9686_ Km app MINK Pass Pass Pass 0.68 PV3810/ SSBK9686_ Km app MKNK1 (MNK1) Pass Pass Pass 0.86 PV6023/ SSBK9686_ Km app MST1R (RON) Pass Pass Pass 0.79 PV4314/ SSBK9686_ Km app MST Pass Pass Pass 0.52 PV3690/ SSBK9686_ Km app MUSK Pass Pass Pass 0.78 PV3834/ SSBK9686_ Km app MYLK2 (skmlck) Pass Pass Pass 0.54 PV3757/ SSBK9686_ Km app NEK Pass Pass Pass 0.78 PV4202/ SSBK9686_ Km app NEK Pass Pass Pass 0.59 PV3360/ SSBK9686_ Km app NEK Pass Pass Pass 0.90 PV4315/ SSBK9686_ Km app NEK Pass Pass Pass 0.83 PV3353/30778 SSBK9686_ Km app NEK Pass Pass Pass 0.92 PV3833/34387 SSBK9686_ Km app NEK Pass Pass Pass 0.79 PV4653/38162 SSBK9686_ Km app NTRK1 (TRKA) Pass Pass Pass 0.84 PV3144/ SSBK9686_ Km app NTRK2 (TRKB) Pass Pass Pass 0.86 PV3616/35706 SSBK9686_ Km app NTRK3 (TRKC) Pass Pass Pass 0.81 PV3617/ SSBK9686_ Km app PAK Pass Pass Pass 0.57 PV3820/35463 SSBK9686_ Km app PAK2 (PAK65) Pass Pass Pass 0.80 PV4565/ SSBK9686_ Km app PAK Pass Pass Pass 0.58 PV3789/34118 SSBK9686_ Km app PAK Pass Pass Pass 0.75 PV4212/ SSBK9686_ Km app PAK Pass Pass Pass 0.54 PV3502/31794 SSBK9686_ Km app PAK7 (KIAA1264) Pass Pass Pass 0.79 PV4405/36846 SSBK9686_ Km app PASK Pass Pass Pass 0.82 PV3972/ SSBK9686_ Km app PDGFRA (PDGFR alpha) Pass Pass Pass 0.82 PV3811/ SSBK9686_ Km app PDGFRA D842V Pass Pass Pass 0.84 PV4203/ SSBK9686_ Km app PDGFRA T674I Pass Pass Pass 0.80 PV3847/35891 SSBK9686_ Km app PDGFRA V561D Pass Pass Pass 0.85 PV4680/ SSBK9686_ Km app PDGFRB (PDGFR beta) Pass Pass Pass 0.94 P3082/27567 SSBK9686_ Km app PDK1 Direct Pass Pass Pass 0.56 P3001/ SSBK9686_ PDK Pass Pass Pass 0.60 P3001/ SSBK9686_ Km app PHKG Pass Pass Pass 0.70 PV3853/ SSBK9686_ Km app PHKG Pass Pass Pass 0.66 PV4555/37321 SSBK9686_ Km app PIM Pass Pass Pass 0.71 PV3503/ SSBK9686_ Km app PIM Pass Pass Pass 0.64 PV3649/32930 SSBK9686_ Km app PKN1 (PRK1) Pass Pass Pass 0.60 PV3790/ SSBK9686_ Km app PLK Pass Pass Pass 0.84 PV3501/ SSBK9686_ Km app PLK Pass Pass Pass 0.86 PV4204/38798 SSBK9686_ Km app PLK Pass Pass Pass 0.64 PV3812/38812 SSBK9686_ Km app PRKACA (PKA) Pass Pass Pass 0.80 P2912/37377 SSBK9686_ Km app PRKCA (PKC alpha) Pass Pass Pass 0.78 P2232/38479 SSBK9686_ Km app PRKCB1 (PKC beta I) Pass Pass Pass 0.59 P2291/ SSBK9686_ Km app PRKCB2 (PKC beta II) Pass Pass Pass 0.56 P2254/ SSBK9686_ Km app PRKCD (PKC delta) Pass Pass Pass 0.55 P2293/39038 SSBK9686_ Km app PRKCE (PKC epsilon) Pass Pass Pass 0.60 P2292/ SSBK9686_ Km app PRKCG (PKC gamma) Pass Pass Pass 0.83 P2233/39126 SSBK9686_ Km app PRKCH (PKC eta) Pass Pass Pass 0.80 P2633/25587 SSBK9686_ Km app PRKCI (PKC iota) Pass Pass Pass 0.88 PV3183/28662 SSBK9686_ Km app PRKCN (PKD3) Pass Pass Pass 0.78 PV3692/ SSBK9686_ Km app PRKCQ (PKC theta) Pass Pass Pass 0.71 P2996/26231 SSBK9686_ Km app PRKCZ (PKC zeta) Pass Pass Pass 0.54 P2273/31602 SSBK9686_ Km app PRKD1 (PKC mu) Pass Pass Pass 0.78 PV3791/34226 SSBK9686_ Km app PRKD2 (PKD2) Pass Pass Pass 0.91 PV3758/34015 SSBK9686_ Km app PRKG Pass Pass Pass 0.81 PV4340/ SSBK9686_ Km app PRKG2 (PKG2) Pass Pass Pass 0.85 PV3973/ SSBK9686_ Km app PRKX Pass Pass Pass 0.78 PV3813/34283 SSBK9686_ Km app PTK2 (FAK) Pass Pass Pass 0.92 PV3832/ SSBK9686_ Km app PTK2B (FAK2) Pass Pass Pass 0.89 PV4567/ SSBK9686_ Km app PTK6 (Brk) Pass Pass Pass 0.90 PV3291/ SSBK9686_ RAF1 (craf) Y340D Y341D Pass Pass Pass 0.88 PV3805/ SSBK9686_ Km app RET V804L Pass Pass Pass 0.93 PV4397/36640 SSBK9686_ Km app RET Y791F Pass Pass Pass 0.90 PV4396/36639 SSBK9686_ Km app RET Pass Pass Pass 0.90 PV3819/ S5
6 SSBK9686_ Km app ROCK Pass Pass Pass 0.82 PV3691/37178 SSBK9686_ Km app ROCK Pass Pass Pass 0.79 PV3759/ SSBK9686_ Km app ROS Pass Pass Pass 0.92 PV3814/ SSBK9686_ Km app RPS6KA1 (RSK1) Pass Pass Pass 0.79 PV3680/ SSBK9686_ Km app RPS6KA2 (RSK3) Pass Pass Pass 0.84 PV3846/34468 SSBK9686_ Km app RPS6KA3 (RSK2) Pass Pass Pass 0.92 PV3323/ SSBK9686_ Km app RPS6KA4 (MSK2) Pass Pass Pass 0.91 PV3782/ SSBK9686_ Km app RPS6KA5 (MSK1) Pass Pass Pass 0.79 PV3681/ SSBK9686_ Km app RPS6KA6 (RSK4) Pass Pass Pass 0.88 PV4557/37496 SSBK9686_ Km app RPS6KB1 (p70s6k) Pass Pass Pass 0.78 PV3815/38944 SSBK9686_ Km app SGK (SGK1) Pass Pass Pass 0.83 PV3818/ SSBK9686_ Km app SGK Pass Pass Pass 0.82 PV3858/ SSBK9686_ Km app SGKL (SGK3) Pass Pass Pass 0.85 PV3859/38954 SSBK9686_ Km app SNF1LK Pass Pass Pass 0.88 PV4792/ SSBK9686_ Km app SRC N Pass Pass Pass 0.84 P2904/21068 SSBK9686_ Km app SRC Pass Pass Pass 0.89 P3044/ SSBK9686_ Km app SRMS (Srm) Pass Pass Pass 0.87 PV4214/ SSBK9686_ Km app SRPK Pass Pass Pass 0.89 PV4215/ SSBK9686_ Km app SRPK Pass Pass Pass 0.54 PV3829/ SSBK9686_ Km app STK22B (TSSK2) Pass Pass Pass 0.80 PV3622/32396 SSBK9686_ Km app STK22D (TSSK1) Pass Pass Pass 0.76 PV3505/ SSBK9686_ Km app STK23 (MSSK1) Pass Pass Pass 0.89 PV3880/ SSBK9686_ Km app STK24 (MST3) Pass Pass Pass 0.54 PV3650/32932 SSBK9686_ Km app STK25 (YSK1) Pass Pass Pass 0.84 PV3657/33163 SSBK9686_ Km app STK3 (MST2) Pass Pass Pass 0.80 PV4805/ SSBK9686_ Km app STK4 (MST1) Pass Pass Pass 0.62 PV3854/38994 SSBK9686_ Km app SYK Pass Pass Pass 0.93 PV3857/ SSBK9686_ Km app TAOK2 (TAO1) Pass Pass Pass 0.67 PV3760/ SSBK9686_ Km app TBK Pass Pass Pass 0.83 PV3504/ SSBK9686_ Km app TEK (Tie2) Pass Pass Pass 0.75 PV3628/34398 SSBK9686_ Km app TXK Pass Pass Pass 0.80 PV5860/ SSBK9686_ Km app TYK Pass Pass Pass 0.90 PV4790/ SSBK9686_ Km app TYRO3 (RSE) Pass Pass Pass 0.67 PV3828/ SSBK9686_ Km app YES Pass Pass Pass 0.84 A15557/ SSBK9686_ Km app ZAP Pass Pass Pass 0.96 P2782/ S6
7 Lantha Life Technologies' SelectScreen Profiling Service: Single Point Results SelectScreen Scientist: Derek Johnston Date: SSBK-LanthaScreen Binding (Paisley UK) Quality Assurance Review: Andrew Heuze Date: Legend Z' Determination Pass < 40% Inhibition 40% - 80% Inhibition 80% Inhibition Project # Compound No. Difference 1X Test Compound Between Data Concentration Kinase Tested % Displacement % Displacement Points Test Compound Interference (nm) Point 1 Point 2 mean Point 1 - Point 2 Donor Acceptor SSBK9686_ ABL1 H396P Pass Pass 0.79 PV6148/ SSBK9686_ ABL1 M351T Pass Pass 0.53 PV6151/ SSBK9686_ ABL1 Q252H Pass Pass 0.94 PV6154/ SSBK9686_ ACVR1 (ALK2) R206H Pass Pass 0.65 PV6232/ SSBK9686_ ACVR1 (ALK2) Pass Pass 0.89 PV4877/ SSBK9686_ ACVR2A Pass Pass 0.73 PV6124/ SSBK9686_ ACVR2B Pass Pass 0.76 PV6049/ SSBK9686_ ACVRL1 (ALK1) Pass Pass 0.58 PV4883/ SSBK9686_ ALK C1156Y Pass Pass 0.94 PV6157/ SSBK9686_ ALK F1174L Pass Pass 0.79 PV6160/ SSBK9686_ ALK L1196M Pass Pass 0.89 PV6166/ SSBK9686_ ALK R1275Q Pass Pass 0.74 PV6169/ SSBK9686_ AMPK (A1/B1/G2) Pass Pass 0.80 PV6238/ SSBK9686_ AMPK (A1/B1/G3) Pass Pass 0.88 PV6241/ SSBK9686_ AMPK (A1/B2/G1) Pass Pass 0.91 PV6244/ SSBK9686_ AMPK (A2/B2/G1) Pass Pass 0.95 PV6247/ SSBK9686_ AMPK (A2/B2/G2) Pass Pass 0.93 PV6250/ SSBK9686_ AXL R499C Pass Pass 0.82 PV6253/ SSBK9686_ BMPR1A (ALK3) Pass Pass 0.81 PV6038/ SSBK9686_ BMPR1B (ALK6) Pass Pass 0.60 PV6235/ SSBK9686_ BMPR Pass Pass 0.81 PV6256/ SSBK9686_ BRAF V599E Pass Pass 0.74 PV3849/ SSBK9686_ BRAF Pass Pass 0.92 PV3848/ SSBK9686_ BRSK Pass Pass 0.74 PV6259/ SSBK9686_ CAMK2G (CaMKII gamma) Pass Pass 0.95 PV6268/ SSBK9686_ CAMKK1 (CAMKKA) Pass Pass 0.79 PV4670/ SSBK9686_ CAMKK2 (CaMKK beta) Pass Pass 0.95 PV4206/35319 SSBK9686_ CASK Pass Pass 0.83 PV6271/ SSBK9686_ CDC7/DBF Pass Pass 0.81 PV6274/ SSBK9686_ CDK1/cyclin A Pass Pass 0.58 PV6280/ SSBK9686_ CDK11 (Inactive) Pass Pass 0.92 PV6283/ SSBK9686_ CDK14 (PFTK1)/cyclin Y Pass Pass 0.71 PV6382/ SSBK9686_ CDK16 (PCTK1)/cyclin Y Pass Pass 0.78 PV6379/ SSBK9686_ CDK2/cyclin A Pass Pass 0.72 PV6289/ SSBK9686_ CDK2/cyclin E Pass Pass 0.75 PV6295/ SSBK9686_ CDK2/cyclin O Pass Pass 0.72 PV6286/ SSBK9686_ CDK3/cyclin E Pass Pass 0.72 PV6298/ SSBK9686_ CDK5 (Inactive) Pass Pass 0.56 PV6301/ SSBK9686_ CDK8/cyclin C Pass Pass 0.91 PV4402/ SSBK9686_ CDK9 (Inactive) Pass Pass 0.73 PV6304/ SSBK9686_ CDK9/cyclin K Pass Pass 0.82 PV4335/35774 SSBK9686_ CLK Pass Pass 0.83 PV3839/ SSBK9686_ DAPK Pass Pass 0.75 PV3614/32159 SSBK9686_ DDR Pass Pass 0.93 PV6047/ SSBK9686_ DDR2 N456S Pass Pass 0.86 PV6172/ SSBK9686_ DDR2 T654M Pass Pass 0.58 PV6175/ SSBK9686_ DDR Pass Pass 0.90 PV3870/ SSBK9686_ DMPK Pass Pass 0.95 PV3784/ SSBK9686_ DYRK Pass Pass 0.57 PV6331/ SSBK9686_ EGFR (ErbB1) d Pass Pass 0.60 PV6178/ SSBK9686_ EIF2AK2 (PKR) Pass Pass 0.57 PV4821/ SSBK9686_ EPHA Pass Pass 0.87 PV3359/30916 SSBK9686_ EPHA Pass Pass 0.72 PV6337/ SSBK9686_ EPHA Pass Pass 0.84 PV3689/33790 SSBK9686_ FGFR1 V561M Pass Pass 0.93 PV6343/ SSBK9686_ FGFR3 G697C Pass Pass 0.57 PV6184/ SSBK9686_ FGFR3 K650M Pass Pass 0.76 PV6187/ SSBK9686_ FLT3 ITD Pass Pass 0.64 PV6190/ SSBK9686_ FYN A Pass Pass 0.80 PV6346/ SSBK9686_ GRK Pass Pass 0.92 PV6352/ SSBK9686_ ICK Pass Pass 0.87 PV6358/ SSBK9686_ KIT A829P Pass Pass 0.89 PV6193/ SSBK9686_ KIT D816H Pass Pass 0.57 PV6196/ SSBK9686_ KIT D816V Pass Pass 0.67 PV6199/ SSBK9686_ KIT D820E Pass Pass 0.89 PV6307/ SSBK9686_ KIT N822K Pass Pass 0.90 PV6310/ SSBK9686_ KIT T670E Pass Pass 0.94 PV6313/ SSBK9686_ KIT V559D T670I Pass Pass 0.93 PV6316/ SSBK9686_ KIT V654A Pass Pass 0.80 PV4132/35129 SSBK9686_ KIT Y823D Pass Pass 0.77 PV6322/ SSBK9686_ LATS Pass Pass 0.80 PV6361/ SSBK9686_ LATS Pass Pass 0.97 PV6364/ SSBK9686_ LIMK Pass Pass 0.89 PV4337/ SSBK9686_ LIMK Pass Pass 0.92 PV3860/ SSBK9686_ MAP2K1 (MEK1) S218D S222D Pass Pass 0.92 P3099/38541 SSBK9686_ MAP2K1 (MEK1) Pass Pass 0.93 PV3303/ SSBK9686_ MAP2K2 (MEK2) Pass Pass 0.95 PV3615/32519 SSBK9686_ MAP2K3 (MEK3) Pass Pass 0.76 PV3662/ SSBK9686_ MAP2K6 (MKK6) S207E T211E Pass Pass 0.85 PV3293/ SSBK9686_ MAP2K6 (MKK6) Pass Pass 0.81 PV3318/ SSBK9686_ MAP3K10 (MLK2) Pass Pass 0.94 PV3877/34554 SSBK9686_ MAP3K11 (MLK3) Pass Pass 0.93 PV3788/ SSBK9686_ MAP3K14 (NIK) Pass Pass 0.90 PV4902/ SSBK9686_ MAP3K2 (MEKK2) Pass Pass 0.58 PV3822/ SSBK9686_ MAP3K3 (MEKK3) Pass Pass 0.85 PV3876/ SSBK9686_ MAP3K5 (ASK1) Pass Pass 0.74 PV3809/ SSBK9686_ MAP3K7/MAP3K7IP1 (TAK1-TAB1) Pass Pass 0.72 PV4394/ SSBK9686_ MAP4K1 (HPK1) Pass Pass 0.79 PV6355/ SSBK9686_ MAP4K3 (GLK) Pass Pass 0.79 PV6349/ SSBK9686_ MAPK10 (JNK3) Pass Pass 0.90 PV4563/ SSBK9686_ MAPK15 (ERK7) Pass Pass 0.93 PV6181/ SSBK9686_ MAPK8 (JNK1) Pass Pass 0.80 PV3319/ SSBK9686_ MAPK9 (JNK2) Pass Pass 0.94 PV3620/32388 SSBK9686_ MERTK (cmer) A708S Pass Pass 0.91 PV6325/ SSBK9686_ MET D1228H Pass Pass 0.90 PV6208/ SSBK9686_ MKNK2 (MNK2) Pass Pass 0.68 PV5607/ SSBK9686_ MLCK (MLCK2) Pass Pass 0.95 PV3835/34028 SSBK9686_ MYLK (MLCK) Pass Pass 0.97 PV4339/36152 SSBK9686_ MYO3B (MYO3 beta) Pass Pass 0.89 PV6367/ SSBK9686_ NLK Pass Pass 0.93 PV4309/35323 SSBK9686_ NUAK Pass Pass 0.68 PV6376/ Z' Kinase Part# / Lot# S7
8 SSBK9686_ PKN2 (PRK2) Pass Pass 0.74 PV3879/ SSBK9686_ PLK Pass Pass 0.82 PV6394/ SSBK9686_ PRKACB (PRKAC beta) Pass Pass 0.79 PV6388/ SSBK9686_ PRKACG (PRKAC gamma) Pass Pass 0.88 PV6391/ SSBK9686_ RAF1 (craf) Y340D Y341D Pass Pass 0.88 PV3805/ SSBK9686_ RET G691S Pass Pass 0.60 PV6214/ SSBK9686_ RET M918T Pass Pass 0.80 PV6217/ SSBK9686_ RET V804M Pass Pass 0.65 PV6223/ SSBK9686_ RIPK Pass Pass 0.94 PV4213/35334 SSBK9686_ RIPK Pass Pass 0.71 PV6397/ SSBK9686_ SIK Pass Pass 0.94 PV6445/ SSBK9686_ SIK Pass Pass 0.70 PV6403/ SSBK9686_ SLK Pass Pass 0.91 PV3830/34390 SSBK9686_ STK16 (PKL12) Pass Pass 0.81 PV4311/36847 SSBK9686_ STK17A (DRAK1) Pass Pass 0.92 PV3783/ SSBK9686_ STK17B (DRAK2) Pass Pass 0.63 PV6328/ SSBK9686_ STK32B (YANK2) Pass Pass 0.93 PV6406/ SSBK9686_ STK32C (YANK3) Pass Pass 0.88 PV6409/ SSBK9686_ STK Pass Pass 0.89 PV4343/ SSBK9686_ STK38 (NDR) Pass Pass 0.54 PV6370/ SSBK9686_ STK38L (NDR2) Pass Pass 0.51 PV6373/ SSBK9686_ STK39 (STLK3) Pass Pass 0.58 PV6412/ SSBK9686_ TAOK Pass Pass 0.86 PV6415/ SSBK9686_ TAOK3 (JIK) Pass Pass 0.71 PV3652/32935 SSBK9686_ TEC Pass Pass 0.91 PV3269/ SSBK9686_ TEK (TIE2) R849W Pass Pass 0.90 PV6226/ SSBK9686_ TEK (TIE2) Y1108F Pass Pass 0.55 PV6229/ SSBK9686_ TESK Pass Pass 0.73 PV6418/ SSBK9686_ TGFBR1 (ALK5) Pass Pass 0.70 PV5837/ SSBK9686_ TGFBR Pass Pass 0.54 PV6122/ SSBK9686_ TLK Pass Pass 0.82 PV6421/ SSBK9686_ TLK Pass Pass 0.93 PV6424/ SSBK9686_ TNIK Pass Pass 0.91 PV6427/ SSBK9686_ TNK2 (ACK) Pass Pass 0.98 PV4807/ SSBK9686_ TTK Pass Pass 0.57 PV3792/ SSBK9686_ ULK Pass Pass 0.95 PV6430/ SSBK9686_ ULK Pass Pass 0.75 PV6433/ SSBK9686_ ULK Pass Pass 0.88 PV6436/ SSBK9686_ WEE Pass Pass 0.78 PV3817/ SSBK9686_ WNK Pass Pass 0.82 PV4341/35976 SSBK9686_ WNK Pass Pass 0.55 PV4342/36047 SSBK9686_ ZAK Pass Pass 0.87 PV3882/34603 S8
9 Adapta Life Technologies' SelectScreen Profiling Service: Single Point Results SelectScreen Scientist: Derek Johnston Date: SSBK-Adapta (Paisley UK) Quality Assurance Review: Catriona MacKay Date: Legend < 40% Inhibition % Conversion Pass 40% - 80% Inhibition Z' Determination Pass 80% Inhibition Project # Compound No. 1X Test Compound Concentration Difference [ATP] Between Data Tested Kinase Tested % Inhibition % Inhibition Points Test Compound Interference (nm) (μm) Point 1 Point 2 mean Point 1 - Point 2 Donor Acceptor SSBK9686_ CAMK1 (CaMK1) Pass Pass 0.51 PV4391/36046 SSBK9686_ Km app CDK7/cyclin H/MNAT Pass Pass 0.81 PV3868/ SSBK9686_ Km app CDK9/cyclin T Pass Pass 0.83 PV4131/ SSBK9686_ Km app CHUK (IKK alpha) Pass Pass 0.68 PV4310/ SSBK9686_ Km app DAPK Pass Pass 0.53 PV3969/32654 SSBK9686_ Km app GSG2 (Haspin) Pass Pass 0.80 PV5708/ SSBK9686_ Km app IRAK Pass Pass 0.62 PV4403/ SSBK9686_ Km app LRRK2 FL Pass Pass 0.82 A15197PT/ SSBK9686_ Km app LRRK2 G2019S FL Pass Pass 0.66 A15200PT/ SSBK9686_ Km app LRRK2 G2019S Pass Pass 0.71 PV4881/ SSBK9686_ Km app LRRK2 I2020T Pass Pass 0.67 PV5854/ SSBK9686_ Km app LRRK2 R1441C Pass Pass 0.78 PV5858/ SSBK9686_ Km app LRRK Pass Pass 0.54 PV4873/ SSBK9686_ Km app NUAK1 (ARK5) Pass Pass 0.78 PV4127/ SSBK9686_ PI4KA (PI4K alpha) Pass Pass 0.60 PV5689/ SSBK9686_ Km app PI4KB (PI4K beta) Pass Pass 0.51 PV5277/ SSBK9686_ Km app PIK3C2A (PI3K-C2 alpha) Pass Pass 0.79 PV5586/ SSBK9686_ PIK3C2B (PI3K-C2 beta) Pass Pass 0.61 PV5374/ SSBK9686_ Km app PIK3C3 (hvps34) Pass Pass 0.61 PV5126/ SSBK9686_ Km app PIK3CA/PIK3R1 (p110 alpha/p85 alpha) Pass Pass 0.55 PV4788/ SSBK9686_ Km app PIK3CB/PIK3R1 (p110 beta/p85 alpha) Pass Pass 0.64 PV6454/52685 SSBK9686_ Km app PIK3CD/PIK3R1 (p110 delta/p85 alpha) Pass Pass 0.60 PV5273/ SSBK9686_ Km app PIK3CG (p110 gamma) Pass Pass 0.80 PV4786/ SSBK9686_ Km app SPHK Pass Pass 0.67 PV5214/ SSBK9686_ SPHK Pass Pass 0.51 PV5216/ Z' Kinase Part# / Lot# S9
10 Supporting Information 3 (Time dependency for CDK12 inhibition) Supplementary Table 1. Time dependency for CDK12 inhibition Compound 0h 1h 2h 5h >10 >10 >10 >10 3 (THZ-531) Staurosporine IC50 values ( M) for each preincubation time were described. Evaluation of time-dependent inhibition of human CDK12 using the LANCE system. Various concentrations of inhibitors in DMSO (50 nl) were dispensed in the assay plate (product # , Greiner Bio-One, Frickenhausen, Germany) using an Access Echo555 liquid handler (Labsite, Sunnyvale, CA). The enzymatic reaction was performed in reaction buffer (25 mm HEPES (ph 7.5), 10 mm MgCl2, 0.01% Tween 20, 1 mm DTT). To evaluate effect of preincubation with inhibitors on CDK12 inhibitory activity, 2.5 μl of 60 nm CDK12 protein was dispensed into each well and incubated for various periods of time at room temperature. Subsequently, the enzymatic reaction was initiated by the addition of 2.5 μl substrate premix (20 μm ATP and 300 nm ULight-4E-BP1 (Thr37/Thr46)), and the plate was incubated for 60 min at room temperature. For the evaluation of CDK12 inhibitory activity of inhibitors without preincubation, 2.5 μl of substrate mixture was added to the plate, followed by the addition of 2.5 μl of 60 nm CDK12 protein and incubation for 60 min at room temperature. The final concentrations of CDK12, ATP, and peptide were 30 nm, 10 μm, and 150 nm, respectively. The enzymatic reaction was terminated by the addition of 5 μl LANCE detection buffer (20 mm S10
11 EDTA (final 10 mm) and 0.30 nm Eu-labeled anti-phospho-eif4e-binding protein 1 (Thr37/46) antibody (final 0.15 nm)). The plate was incubated for 60 min at room temperature and the time-resolved fluorescence was monitored by using an EnVision plate reader. The solution in each well was excited with a laser (λ = 337 nm) reflected by a dichroic mirror (D400/D630 (PerkinElmer)), and the fluorescence signals from Eu and ULight were detected by using two emission filters (Cy5 620 (PerkinElmer) for Eu and APC 665 (PerkinElmer) for ULight). The values for the 0% and 100% controls were the signals obtained in the presence and absence of CDK12 protein, respectively. The inhibition at each inhibitor concentration was determined in quadruplicate. The IC50 values were derived from the fitting a sigmoidal dose-response curve to a plot of the assay readout over the inhibitor concentration. All fits were computed by GraphPad Prism. S11
12 Supporting Information 4 (Co-crystal structure of 29 and CDK12) Protein expression and purification. The cdnas encoding human CDK12 and CycK (encompassing residues and 1 267, respectively) were amplified by the polymerase chain reaction and subcloned into the plasmid pfastbacdual (Invitrogen). The CDK12 coding sequence was inserted after the polyhedron promoter (PpH), whereas CycK was placed behind the p10 promoter (Pp10). To enable the purification of the complex, the coding sequences were each engineered with an N-terminal Glutathione-S-transferase (GST) affinity tag that includes a recognition sequence for the TEV protease for cleavage and removal of GST fusion partner protein. In addition, the cdna encoding S. cerevisiae CDK-activating kinase (CAK1p) encompassing residues was amplified and subcloned into the plasmid pfastbac. The CAK1p construct was engineered with an N-terminal 6X-His affinity tag and a TEV recognition sequence to facilitate purification in the event that the isolated CAK1p was required for in vitro experiments. After construction, the cloned inserts were verified by DNA sequence analysis. Recombinant viruses were generated using the Bac-to-Bac baculovirus expression system (Invitrogen). High-titer viruses encoding CDK12/CycK and CAK1p were mixed in a 1:1 ratio and Hi5 cells were infected to express the recombinant proteins. The CAK1p was included during expression as the phosphorylation of CDK12 was reported to facilitate crystallization. 1 The infected Hi5 cells were harvested 66 h after the addition of the viruses. A cell pellet generated from a 10 L culture of Hi5 cells was homogenized in 1 L of lysate buffer (25 mm HEPES (ph 7.6), 0.5 M NaCl, 0.25 mm TCEP, and 5% glycerol; all chemicals were purchased from Sigma-Aldrich) supplemented with protease inhibitors (Roche). The lysate was clarified by centrifugation and the resulting supernatant was affinity purified by using glutathione sepharose (GE Healthcare). The glutathione affinity column was washed with 50 volumes of lysate buffer S12
13 in the absence of protease inhibitors. To remove the GST-tag, the column was incubated with TEV protease overnight during an on-column cleavage step and the following day the CDK12/CycK protein complex was eluted with buffer (25 mm HEPES ph 7.6, 0.4 M NaCl, 0.5 mm TCEP, 5% glycerol). The complex was further purified in this buffer by passage through a size exclusion column (Superdex 200, GE Healthcare). The peak fractions from size exclusion chromatography that contained the CDK12/CycK heterodimeric complex were pooled and stored in aliquots at 80 C. Analysis of the purified complex by mass spectrometry demonstrated that CDK12 was phosphorylated (data not shown). Crystallography. Prior to setting up crystallization trials, a CDK12/CycK-compound ternary complex was generated by the addition of a macrocyclic ATP-competitive compound (the identity of this tool compound is not disclosed) followed by co-concentration using an Amicon concentrator (EMD-Millipore) to produce a final concentration of 12 mg/ml. Crystallization was performed using the hanging drop vapor diffusion method with a reservoir solution composed of 0.1 M HEPES (ph 6.5), 18% PEG 3350, 0.2 M MgCl2, and 1 mm sarcosine (the additive sarcosine and PEG3350 solution were purchased from Hampton Research). The crystals were grown at 25 C and reaching their maximum size within 1 month. The crystals of CDK12/CycK were backsoaked with mother liquor supplemented with 2 mm compound 29 by transferring the crystals into fresh compound/mother liquor solution once per day for five consecutive days. After the soaking regimen, the crystals were cryo-protected by rapid transfer into a reservoir solution supplemented with 25% ethylene glycol and rapidly flash-cooled in ALS-style pucks that were submerged in liquid nitrogen. The diffraction data were collected at Advanced Light Source beamline (Lawrence Berkeley s National Laboratory, Berkeley, CA) and processed using the HKL2000 software package. 2 The structure was solved by molecular replacement with S13
14 Phaser 3 using stepwise searches with the coordinates of CDK12 followed by CycK (both models derived from PDB code:4nst). The graphics program COOT 4 was used for model building and refinement was performed with REFMAC5. 5 Phaser and REFMAC5 are distributed as part of CCP4. 6 Structural validation was performed by using Molprobity. 7 Refinement statistics are reported in Supplementary Table 2 and an image of the omit electron density for the ligand is shown in Supplementary Figure 2. S14
15 Supplementary Table 2. X-ray data collection and refinement statistics. X-Ray Structure CDK12/CycK-29 /PDB CODE: 6CKX Data collection Resolution range (Å) ( ) Space group P Cell dimensions: a, b, c (Å) , , α, β, γ (º) , , Rsym <I / σi> Completeness (%) Redundancy Structure Refinement Resolution (Å) No. reflections No. reflections Rfree test set Rwork / Rfree No. atoms: Protein Ligand Mg Ions Waters Mean B Value (Å 2 ) Protein B Value (Å 2 ) Ligand B Value (Å 2 ) Mg Ions B Value (Å 2 ) Water B Value (Å 2 ) RMS Bond lengths (Å) RMS Bond angles (º) Ramachandran statistics: Favored (%) 10.7 (100) 13.9 (1.05) 99.9 (99.8) 6.9 (6.7) ,375 1, /25.1 8, S15
16 Outliers (%) 0.19 *Values in parentheses are for highest-resolution shell. Supplementary Figure 2. Omit electron density Fo-Fc map (rendered as mesh) was generated by omitting atoms corresponding to the compound (displayed as green sticks). Map is contoured at ± 3.0 σ. Figure was created using Pymol. S16
17 Supporting Information 5 (Single crystal X-ray structure analyses) All measurements were made on a Rigaku R-AXIS RAPID-191R or a Rigaku XtaLAB P200 diffractometer using graphite monochromated Cu-Kα radiation. The structure was solved by direct methods with SIR and was refined using full-matrix least-squares on F 2 with SHELXL All non-h atoms were refined with anisotropic displacement parameters. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via Supplementary Figure 3. ORTEP of 1, thermal ellipsoids are drawn at 50% probability. Crystal data for 1 (CCDC ): C21H22N6O2S, MW = ; crystal size, 0.30 x 0.08 x 0.06 mm; colorless, block; monoclinic, space group P21/n, a = (19) Å, b = (5) Å, c = (6) Å, α = γ = 90, β = (6), V = (10) Å 3, Z = 4, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.073, wr2 = S17
18 Supplementary Figure 4. ORTEP of 16, thermal ellipsoids are drawn at 50% probability. Crystal data for 16 (CCDC ): C22H22N6O, MW = ; crystal size, 0.12 x 0.10 x 0.04 mm; colorless, block; monoclinic, space group P21/n, a = (14) Å, b = (14) Å, c = (3) Å, α = γ = 90, β = (14), V = (5) Å 3, Z = 4, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.045, wr2 = Supplementary Figure 5. ORTEP of 21, thermal ellipsoids are drawn at 50% probability. Crystal data for 21 (CCDC ): C24H26N6O 0.4 H2O, MW = ; crystal size, 0.21 x 0.11 x 0.06 mm; colorless, platelet; triclinic, space group P-1, a = (4) Å, b = (5) S18
19 Å, c = (5) Å, α = (7), β = (8), γ = (8), V = (13) Å 3, Z = 2, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.130, wr2 = Supplementary Figure 6. ORTEP of 22, thermal ellipsoids are drawn at 50% probability. Crystal data for 22 (CCDC ): C23H26N6O2S, MW = ; crystal size, 0.34 x 0.23 x 0.06 mm; colorless, block; orthorhombic, space group P212121, a = (7) Å, b = (8) Å, c = (15) Å, α = β = γ = 90, V = (3) Å 3, Z = 4, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.026, wr2 = Supplementary Figure 7. ORTEP of 23, thermal ellipsoids are drawn at 50% probability. S19
20 Crystal data for 23 (CCDC ): C31H32N6O 1.0 C3H8O, MW = ; crystal size, 0.14 x 0.04 x 0.04 mm; colorless, block; triclinic, space group P-1, a = (3) Å, b = (3) Å, c = (5) Å, α = (2), β = (3), γ = (2), V = (8) Å 3, Z = 2, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.046, wr2 = Supplementary Figure 8. ORTEP of 25, thermal ellipsoids are drawn at 50% probability. Crystal data for 25 (CCDC ): C30H30N6O2 C3H8O, MW = ; crystal size, 0.18 x 0.11 x 0.05 mm; colorless, block; triclinic, space group P-1, a = (19) Å, b = (2) Å, c = (3) Å, α = (16), β = (16), γ = (17), V = (5) Å 3, Z = 2, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.041, wr2 = S20
21 Supplementary Figure 9. ORTEP of 27, thermal ellipsoids are drawn at 50% probability. Crystal data for 27 (CCDC ): C30H31N7O, MW = ; crystal size, 0.13 x 0.11 x 0.07 mm; colorless, platelet; triclinic, space group P-1, a = (2) Å, b = (3) Å, c = (3) Å, α = (6), β = (5), γ = (5), V = (8) Å 3, Z = 2, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.069, wr2 = Supplementary Figure 10. ORTEP of 33, thermal ellipsoids are drawn at 50% probability. Crystal data for 33 (CCDC ): C24H26N6O, MW = ; crystal size, 0.21 x 0.04 x 0.04 mm; colorless, needle; monoclinic, space group P21/c, a = (16) Å, b = S21
22 (12) Å, c = (2) Å, α = γ = 90, β = (12), V = (8) Å 3, Z = 8, Dx = g/cm 3, T = 100 K, μ = mm -1, λ = Å, R1 = 0.037, wr2 = S22
23 References 1. Bösken C. A., Farnung L., Hintermair C., Merzel Schachter M., Vogel-Bachmayr K., Blazek D., Anand K., Fisher RP., Eick D., Geyer M. The structure and substrate specificity of human Cdk12/Cyclin K. Nat. Commun. 2014, 5, Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, McCoy, A. J., Grosse-Kunstleve, R.W., Adams, P. D., Winn, M. D., Storoni, L. C., Read, R. J. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, Emsley, P., Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, Collaborative Computing Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, Burla, M. C.; Caliandro, R.; Camalli, M.; Carrozzini, B.; Cascarano, G. L.; De Caro, L.; Giacovazzo, C.; Polidori, G.; Siliqi, D.; Spagna, R. IL MILIONE: a suite of computer programs for crystal structure solution of proteins. J. Appl. Crystallogr. 2007, 40, Sheldrick, G. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, S23
SelectScreen Kinase Profiling Service Activity Assay Selectivity Data
 BAY-43-9006 BIRB-796 BMS-354825 Gleevec Go-6983 H-89 Iressa NH125 PI-103 PP2 (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) ABL1
BAY-43-9006 BIRB-796 BMS-354825 Gleevec Go-6983 H-89 Iressa NH125 PI-103 PP2 (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) (10,000 nm) ABL1
Supporting Information
 Supporting Information The design of potent and selective inhibitors to overcome clinical ALK mutations resistant to crizotinib Qinhua Huang,* Ted W. Johnson,* Simon Bailey, Alexei Brooun, Kevin D. Bunker,
Supporting Information The design of potent and selective inhibitors to overcome clinical ALK mutations resistant to crizotinib Qinhua Huang,* Ted W. Johnson,* Simon Bailey, Alexei Brooun, Kevin D. Bunker,
Development of highly potent and selective diaminothiazole inhibitors of. cyclin-dependent kinases
 Supporting Information Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases Ernst Schonbrunn a*, Stephane Betzi a, Riazul Alam a, Mathew P. Martin a, Andreas
Supporting Information Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases Ernst Schonbrunn a*, Stephane Betzi a, Riazul Alam a, Mathew P. Martin a, Andreas
RET SRC WEE1 JAK2 MAP2K1 PKMYT1 ERBB2 BMX MET PTK2 FGR MAPK3 MAP2K2 ABL1 MAP2K4 MAP2K6 MAP3K6 MAP3K5 CDK5 ALK PTK2 MAPK14 LCK HIPK2
 4 SYK MAPK9 6 SKMel28.Replicate.1 Kinome 4 MAPK9 DYRK2 SKMel28.Replicate.1 ActLoop MAP3K6 SKMel28.Replicate.1 PSP SRMS SKMel28.Replicate.1 NWK 0 10 20 30 40 50 0 10 20 30 40 50 0 20 40 60 80 100 0 10 20
4 SYK MAPK9 6 SKMel28.Replicate.1 Kinome 4 MAPK9 DYRK2 SKMel28.Replicate.1 ActLoop MAP3K6 SKMel28.Replicate.1 PSP SRMS SKMel28.Replicate.1 NWK 0 10 20 30 40 50 0 10 20 30 40 50 0 20 40 60 80 100 0 10 20
Supporting Information
 Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
A cell-based screening system for RNA Polymerase I inhibitors
 Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Supporting Information
 Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supplementary Table 1. Cystometric parameters in sham-operated wild type and Trpv4 -/- rats during saline infusion and
 WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
The role of aristolochene synthase in diphosphate activation
 Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Construction of a cube given with its centre and a sideline
 Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and
 1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo
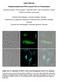
 Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Supporting Information
 Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
NYOMÁSOS ÖNTÉS KÖZBEN ÉBREDŐ NYOMÁSVISZONYOK MÉRÉTECHNOLÓGIAI TERVEZÉSE DEVELOPMENT OF CAVITY PRESSURE MEASUREMENT FOR HIGH PRESURE DIE CASTING
 Anyagmérnöki Tudományok, 39/1 (2016) pp. 82 86. NYOMÁSOS ÖNTÉS KÖZBEN ÉBREDŐ NYOMÁSVISZONYOK MÉRÉTECHNOLÓGIAI TERVEZÉSE DEVELOPMENT OF CAVITY PRESSURE MEASUREMENT FOR HIGH PRESURE DIE CASTING LEDNICZKY
Anyagmérnöki Tudományok, 39/1 (2016) pp. 82 86. NYOMÁSOS ÖNTÉS KÖZBEN ÉBREDŐ NYOMÁSVISZONYOK MÉRÉTECHNOLÓGIAI TERVEZÉSE DEVELOPMENT OF CAVITY PRESSURE MEASUREMENT FOR HIGH PRESURE DIE CASTING LEDNICZKY
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet Nonparametric Tests
 Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Computer Architecture
 Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
KN-CP50. MANUAL (p. 2) Digital compass. ANLEITUNG (s. 4) Digitaler Kompass. GEBRUIKSAANWIJZING (p. 10) Digitaal kompas
 KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
Cluster Analysis. Potyó László
 Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Suppl. Materials. Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids. Germany
 Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Eredeti gyógyszerkutatás. ELTE TTK vegyészhallgatók számára Dr Arányi Péter 2009 március, 4.ea. Szerkezet optimalizálás (I.)
 Eredeti gyógyszerkutatás ELTE TTK vegyészhallgatók számára Dr Arányi Péter 2009 március, 4.ea. Szerkezet optimalizálás (I.) 1 How do we proceed? New target Internal Literature Validation Selected Target
Eredeti gyógyszerkutatás ELTE TTK vegyészhallgatók számára Dr Arányi Péter 2009 március, 4.ea. Szerkezet optimalizálás (I.) 1 How do we proceed? New target Internal Literature Validation Selected Target
- Supplementary Data
 1 Open Access Asian Australas. J. Anim. Sci. Vol. 29, No. 3 : 321-326 March 2016 http://dx.doi.org/10.5713/ajas.15.0331 www.ajas.info pissn 1011-2367 eissn 1976-5517 Analysis of Swine Leukocyte Antigen
1 Open Access Asian Australas. J. Anim. Sci. Vol. 29, No. 3 : 321-326 March 2016 http://dx.doi.org/10.5713/ajas.15.0331 www.ajas.info pissn 1011-2367 eissn 1976-5517 Analysis of Swine Leukocyte Antigen
First experiences with Gd fuel assemblies in. Tamás Parkó, Botond Beliczai AER Symposium 2009.09.21 25.
 First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
Mapping Sequencing Reads to a Reference Genome
 Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
A évi fizikai Nobel-díj
 A 2012. évi fizikai Nobel-díj "for ground-breaking experimental methods that enable measuring and manipulation of individual quantum systems" Serge Haroche David Wineland Ecole Normale Superieure, Párizs
A 2012. évi fizikai Nobel-díj "for ground-breaking experimental methods that enable measuring and manipulation of individual quantum systems" Serge Haroche David Wineland Ecole Normale Superieure, Párizs
Expression analysis of PIN genes in root tips and nodules of Lotus japonicus
 Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
PETER PAZMANY CATHOLIC UNIVERSITY Consortium members SEMMELWEIS UNIVERSITY, DIALOG CAMPUS PUBLISHER
 SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
University of Bristol - Explore Bristol Research
 Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Nonparametric Tests. Petra Petrovics.
 Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
Correlation & Linear Regression in SPSS
 Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092)
 Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092) www.zoolog.hu Dr. Dombos Miklós Tudományos főmunkatárs MTA ATK TAKI Innovative Real-time Monitoring and Pest control
Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092) www.zoolog.hu Dr. Dombos Miklós Tudományos főmunkatárs MTA ATK TAKI Innovative Real-time Monitoring and Pest control
vancomycin CFSL cefepime CFPM cefozopran CZOP 4 cephem cefpirome CPR cefoselis Inhibitory Concentration FIC index
 vancomycin cephem 1a 2 1 1 2 a : 15 4 15 15 9 5 1999 2 methicillin MRSA12 vancomycinvcm 4 cephem cefpiromecprcefoseliscfslcefepimecfpmcefozopranczop VCM CPR 12 77 75.5CFSL 82.4CFPM 81 79.4 CZOP 76 74.5
vancomycin cephem 1a 2 1 1 2 a : 15 4 15 15 9 5 1999 2 methicillin MRSA12 vancomycinvcm 4 cephem cefpiromecprcefoseliscfslcefepimecfpmcefozopranczop VCM CPR 12 77 75.5CFSL 82.4CFPM 81 79.4 CZOP 76 74.5
SZOFTVEREK A SORBANÁLLÁSI ELMÉLET OKTATÁSÁBAN
 SZOFTVEREK A SORBANÁLLÁSI ELMÉLET OKTATÁSÁBAN Almási Béla, almasi@math.klte.hu Sztrik János, jsztrik@math.klte.hu KLTE Matematikai és Informatikai Intézet Abstract This paper gives a short review on software
SZOFTVEREK A SORBANÁLLÁSI ELMÉLET OKTATÁSÁBAN Almási Béla, almasi@math.klte.hu Sztrik János, jsztrik@math.klte.hu KLTE Matematikai és Informatikai Intézet Abstract This paper gives a short review on software
Mikroszerkezet Krisztallitonként Tömbi Polikristályos Mintában
 Mikroszerkezet Krisztallitonként Tömbi Polikristályos Mintában Ribárik Gábor, Zilahi Gyula és Ungár Tamás Anyagfizikai Tanszék TAMOP Szeminárium, Visegrád 2012, január 18-20. Diffrakciós vonalak kiszélesedése
Mikroszerkezet Krisztallitonként Tömbi Polikristályos Mintában Ribárik Gábor, Zilahi Gyula és Ungár Tamás Anyagfizikai Tanszék TAMOP Szeminárium, Visegrád 2012, január 18-20. Diffrakciós vonalak kiszélesedése
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites
 Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
pjnc-pgluc Codon-optimized Gaussia luciferase (pgluc) Vector backbone: JM83 pjnc-pgluc is resistant to ampicillin and neomycin High or low copy:
 pjnc-pgluc Gene/Insert name: Codon-optimized Gaussia luciferase (pgluc) Vector backbone: pcmv-jnc Vector type: Mammalian cells Backbone size w/o insert (bp): 5375 Bacterial resistance: Ampicillin and neomycin
pjnc-pgluc Gene/Insert name: Codon-optimized Gaussia luciferase (pgluc) Vector backbone: pcmv-jnc Vector type: Mammalian cells Backbone size w/o insert (bp): 5375 Bacterial resistance: Ampicillin and neomycin
General information for the participants of the GTG Budapest, 2017 meeting
 General information for the participants of the GTG Budapest, 2017 meeting Currency is Hungarian Forint (HUF). 1 EUR 310 HUF, 1000 HUF 3.20 EUR. Climate is continental, which means cold and dry in February
General information for the participants of the GTG Budapest, 2017 meeting Currency is Hungarian Forint (HUF). 1 EUR 310 HUF, 1000 HUF 3.20 EUR. Climate is continental, which means cold and dry in February
Egyensúlyba hozza olajos bőrét. 0 % PARABEn & PHEnOXYETHAnOL PÓRUSÖSSZEHÚZÓ DERMATOLÓGIAILAG TESZTELT VÁSÁRÓLI FÜZET
 Újdonság ALGOPURE Egyensúlyba hozza olajos bőrét 0 % PARABEn & PHEnOXYETHAnOL PÓRUSÖSSZEHÚZÓ DERMATOLÓGIAILAG TESZTELT VÁSÁRÓLI FÜZET BŐR PROBLÉMÁJA Kombinált és Zsíros bőr egyensúlyának hiánya 1 Túlzott
Újdonság ALGOPURE Egyensúlyba hozza olajos bőrét 0 % PARABEn & PHEnOXYETHAnOL PÓRUSÖSSZEHÚZÓ DERMATOLÓGIAILAG TESZTELT VÁSÁRÓLI FÜZET BŐR PROBLÉMÁJA Kombinált és Zsíros bőr egyensúlyának hiánya 1 Túlzott
Feloldóképesség 2009.12.08. Mikroszkópos módszerek. DIC mikroszkópia. Fáziskontraszt mikroszkópia. Barkó Szilvia A MIKROSZKÓPIA RÖVID TÖRTÉNETE
 A MIKROSZKÓPIA RÖVID TÖRTÉNETE Mikroszkópos módszerek Barkó Szilvia 1667: Robert Hooke cellulákat ír le parafában összetett mikroszkóp segítségével. 1674: Antony van Leeuwenhoek élő mikróbákat figyel meg
A MIKROSZKÓPIA RÖVID TÖRTÉNETE Mikroszkópos módszerek Barkó Szilvia 1667: Robert Hooke cellulákat ír le parafában összetett mikroszkóp segítségével. 1674: Antony van Leeuwenhoek élő mikróbákat figyel meg
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing MFMER slide-1
 Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes
 Stem Cell Reports, Volume 2 Supplemental Information Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes Yuko
Stem Cell Reports, Volume 2 Supplemental Information Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes Yuko
Flowering time. Col C24 Cvi C24xCol C24xCvi ColxCvi
 Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
STUDENT LOGBOOK. 1 week general practice course for the 6 th year medical students SEMMELWEIS EGYETEM. Name of the student:
 STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
Statistical Inference
 Petra Petrovics Statistical Inference 1 st lecture Descriptive Statistics Inferential - it is concerned only with collecting and describing data Population - it is used when tentative conclusions about
Petra Petrovics Statistical Inference 1 st lecture Descriptive Statistics Inferential - it is concerned only with collecting and describing data Population - it is used when tentative conclusions about
Supplemental Information. Investigation of Penicillin Binding Protein. (PBP)-like Peptide Cyclase and Hydrolase
 Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
Akt1 Akt kinase activity Creb signaling CCTTACAGCCCTCAAGTACTCATTC GGCGTACTCCATGACAAAGCA Arc Actin binding
 Additional File 1 (Table S1): Description and primer sequences of 80 candidate genes tested as potential synaptic plasticity formation genes in qrt-pcr array. Abbreviations: LTP: long-term potentiation,
Additional File 1 (Table S1): Description and primer sequences of 80 candidate genes tested as potential synaptic plasticity formation genes in qrt-pcr array. Abbreviations: LTP: long-term potentiation,
AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN
 Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
Modular Optimization of Hemicellulose-utilizing Pathway in. Corynebacterium glutamicum for Consolidated Bioprocessing of
 [Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
[Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
Széchenyi István Egyetem www.sze.hu/~herno
 Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
Új hálózati megoldások Gbit xdsl technológiával
 Új hálózati megoldások Gbit xdsl technológiával ITU szabványok és piaci termékek Mérnöki Kamara előadás 2019. február 18. Takács György Az ITU-T 15. Tanulmányi Bizottság Q4 kérdése VDSL2, G.fast, G.mgfast
Új hálózati megoldások Gbit xdsl technológiával ITU szabványok és piaci termékek Mérnöki Kamara előadás 2019. február 18. Takács György Az ITU-T 15. Tanulmányi Bizottság Q4 kérdése VDSL2, G.fast, G.mgfast
A templát reakciók. Template Reactions
 A templát reakciók Template Reactions ifj. Dr. VÁRHELYI Csaba 1, Dr. PKL György 2, Dr. LIPTAY György 2, Dr. MAJDIK Kornélia 1, Dr. FARKAS György 1, Dr. VÁRHELYI Csaba 1 1 Babeş-Bolyai Tudományegyetem,
A templát reakciók Template Reactions ifj. Dr. VÁRHELYI Csaba 1, Dr. PKL György 2, Dr. LIPTAY György 2, Dr. MAJDIK Kornélia 1, Dr. FARKAS György 1, Dr. VÁRHELYI Csaba 1 1 Babeş-Bolyai Tudományegyetem,
IP/09/473. Brüsszel, 2009. március 25
 IP/09/473 Brüsszel, 2009. március 25 A mobiltelefon-használat nő, míg a fogyasztói árak csökkennek: a Bizottság jelentése szerint az európai távközlési ágazat ellenáll a gazdasági lassulásnak 2008-ban
IP/09/473 Brüsszel, 2009. március 25 A mobiltelefon-használat nő, míg a fogyasztói árak csökkennek: a Bizottság jelentése szerint az európai távközlési ágazat ellenáll a gazdasági lassulásnak 2008-ban
EN United in diversity EN A8-0206/419. Amendment
 22.3.2019 A8-0206/419 419 Article 2 paragraph 4 point a point i (i) the identity of the road transport operator; (i) the identity of the road transport operator by means of its intra-community tax identification
22.3.2019 A8-0206/419 419 Article 2 paragraph 4 point a point i (i) the identity of the road transport operator; (i) the identity of the road transport operator by means of its intra-community tax identification
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon
 Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Correlation & Linear Regression in SPSS
 Correlation & Linear Regression in SPSS Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise 1 - Correlation File / Open
Correlation & Linear Regression in SPSS Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise 1 - Correlation File / Open
Jelátviteli uatk és daganatképződés
 Jelátviteli uatk és daganatképződés Tímár József Semmelweis Egyetem, Klinikai Központ 2.sz. Patológiai Intézet MTA-SE Molekuláris Onkológia Kutatócsoport, Budapest A HER-2 genetikája emberi daganatokban
Jelátviteli uatk és daganatképződés Tímár József Semmelweis Egyetem, Klinikai Központ 2.sz. Patológiai Intézet MTA-SE Molekuláris Onkológia Kutatócsoport, Budapest A HER-2 genetikája emberi daganatokban
Using the CW-Net in a user defined IP network
 Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
CONCERTO COMMUNITIES IN EU DEALING WITH OPTIMAL THERMAL AND ELECTRICAL EFFICIENCY OF BUILDINGS AND DISTRICTS, BASED ON MICROGRIDS. WP 5 Del 5.
 CONCERTO COMMUNITIES IN EU DEALING WITH OPTIMAL THERMAL AND ELECTRICAL EFFICIENCY OF BUILDINGS AND DISTRICTS, BASED ON MICROGRIDS WP 5 Del 5.14 1 st period Szentendre Papers and articles in specialist
CONCERTO COMMUNITIES IN EU DEALING WITH OPTIMAL THERMAL AND ELECTRICAL EFFICIENCY OF BUILDINGS AND DISTRICTS, BASED ON MICROGRIDS WP 5 Del 5.14 1 st period Szentendre Papers and articles in specialist
A rosszindulatú daganatos halálozás változása 1975 és 2001 között Magyarországon
 A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
On The Number Of Slim Semimodular Lattices
 On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
Planetary Nebulae, PN PNe PN
 2 2 e-mail: masaaki@oao.nao.ac.jp 719 0232 3037 5 Subaru Telescope, National Astronomical Observatory of Japan, 650 North A ohoku Place, Hilo, Hawaii 96720, U.S.A. e-mail: tajitsu@subaru.naoj.org 397 0101
2 2 e-mail: masaaki@oao.nao.ac.jp 719 0232 3037 5 Subaru Telescope, National Astronomical Observatory of Japan, 650 North A ohoku Place, Hilo, Hawaii 96720, U.S.A. e-mail: tajitsu@subaru.naoj.org 397 0101
Lexington Public Schools 146 Maple Street Lexington, Massachusetts 02420
 146 Maple Street Lexington, Massachusetts 02420 Surplus Printing Equipment For Sale Key Dates/Times: Item Date Time Location Release of Bid 10/23/2014 11:00 a.m. http://lps.lexingtonma.org (under Quick
146 Maple Street Lexington, Massachusetts 02420 Surplus Printing Equipment For Sale Key Dates/Times: Item Date Time Location Release of Bid 10/23/2014 11:00 a.m. http://lps.lexingtonma.org (under Quick
DG(SANCO)/2012-6290-MR
 1 Ensure official controls of food contaminants across the whole food chain in order to monitor the compliance with the requirements of Regulation (EC) No 1881/2006 in all food establishments, including
1 Ensure official controls of food contaminants across the whole food chain in order to monitor the compliance with the requirements of Regulation (EC) No 1881/2006 in all food establishments, including
CLUSTALW Multiple Sequence Alignment
 Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali. Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang*
 Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Effect of the different parameters to the surface roughness in freeform surface milling
 19 November 0, Budapest Effect of the different parameters to the surface roughness in freeform surface milling Balázs MIKÓ Óbuda University 1 Abstract Effect of the different parameters to the surface
19 November 0, Budapest Effect of the different parameters to the surface roughness in freeform surface milling Balázs MIKÓ Óbuda University 1 Abstract Effect of the different parameters to the surface
Phenotype. Genotype. It is like any other experiment! What is a bioinformatics experiment? Remember the Goal. Infectious Disease Paradigm
 It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
Eladni könnyedén? Oracle Sales Cloud. Horváth Tünde Principal Sales Consultant 2014. március 23.
 Eladni könnyedén? Oracle Sales Cloud Horváth Tünde Principal Sales Consultant 2014. március 23. Oracle Confidential Internal/Restricted/Highly Restricted Safe Harbor Statement The following is intended
Eladni könnyedén? Oracle Sales Cloud Horváth Tünde Principal Sales Consultant 2014. március 23. Oracle Confidential Internal/Restricted/Highly Restricted Safe Harbor Statement The following is intended
White Paper. Grounding Patch Panels
 White Paper Grounding Patch Panels Tartalom 1. Bevezető... 1 2. A földelés jelentőssége... 3 3. AC elosztó rendszer... 3 4. Földelési rendszerek... 3 4.1. Fa... 3 4.2. Háló... 5 5. Patch panel földelési
White Paper Grounding Patch Panels Tartalom 1. Bevezető... 1 2. A földelés jelentőssége... 3 3. AC elosztó rendszer... 3 4. Földelési rendszerek... 3 4.1. Fa... 3 4.2. Háló... 5 5. Patch panel földelési
(NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV
 Kommunikációs rendszerek programozása (NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV (5. mérés) SIP telefonközpont készítése Trixbox-szal 1 Mérés helye: Széchenyi István Egyetem, L-1/7 laboratórium, 9026 Győr, Egyetem
Kommunikációs rendszerek programozása (NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV (5. mérés) SIP telefonközpont készítése Trixbox-szal 1 Mérés helye: Széchenyi István Egyetem, L-1/7 laboratórium, 9026 Győr, Egyetem
Genome 373: Hidden Markov Models I. Doug Fowler
 Genome 373: Hidden Markov Models I Doug Fowler Review From Gene Prediction I transcriptional start site G open reading frame transcriptional termination site promoter 5 untranslated region 3 untranslated
Genome 373: Hidden Markov Models I Doug Fowler Review From Gene Prediction I transcriptional start site G open reading frame transcriptional termination site promoter 5 untranslated region 3 untranslated
HTS (High Throughput Screening) Lehetőségek a Debreceni Egyetemen. DE OEC Orvosi Vegytani Intézet
 HTS in HTS (High Throughput Screening) Lehetőségek a Debreceni Egyetemen Virág László DE OEC Orvosi Vegytani Intézet ET III. emelet BIO MMKK DE-BIK Mi a HTS? Robotizált mintamozgatási, műszeres mérési
HTS in HTS (High Throughput Screening) Lehetőségek a Debreceni Egyetemen Virág László DE OEC Orvosi Vegytani Intézet ET III. emelet BIO MMKK DE-BIK Mi a HTS? Robotizált mintamozgatási, műszeres mérési
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Correlation & Linear. Petra Petrovics.
 Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella
 A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella Országos Meteorológiai Szolgálat Hungarian Meteorological Service KRITéR
A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella Országos Meteorológiai Szolgálat Hungarian Meteorological Service KRITéR
Összefoglalás. Summary. Bevezetés
 A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel
 Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel /Study of Solvent Gradient by Simulated Moving Bed Preparative Liqiud Chpomatography Technology/ 1 Nagy
Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel /Study of Solvent Gradient by Simulated Moving Bed Preparative Liqiud Chpomatography Technology/ 1 Nagy
± ± ± ƒ ± ± ± ± ± ± ± ƒ. ± ± ƒ ± ± ± ± ƒ. ± ± ± ± ƒ
 ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ç å ± ƒ ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ä ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ±
± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ç å ± ƒ ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ä ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ±
 Társadalmi-gazdasági szempontok Az ipari termelési folyamatok kedvezőbbé tétele és az ipari együttműködési láncok sűrűsége pozitív társadalmi és gazdasági eredmények létrejöttéhez is hozzájárul. A társadalmi
Társadalmi-gazdasági szempontok Az ipari termelési folyamatok kedvezőbbé tétele és az ipari együttműködési láncok sűrűsége pozitív társadalmi és gazdasági eredmények létrejöttéhez is hozzájárul. A társadalmi
Computational Neuroscience
 Computational Neuroscience Zoltán Somogyvári senior research fellow KFKI Research Institute for Particle and Nuclear Physics Supporting materials: http://www.kfki.hu/~soma/bscs/ BSCS 2010 Lengyel Máté:
Computational Neuroscience Zoltán Somogyvári senior research fellow KFKI Research Institute for Particle and Nuclear Physics Supporting materials: http://www.kfki.hu/~soma/bscs/ BSCS 2010 Lengyel Máté:
DANS és Narcis. Burmeister Erzsébet. HUNOR találkozó, Budapest 2013. március 13.
 DANS és Narcis Burmeister Erzsébet HUNOR találkozó, Budapest 2013. március 13. DANS DANS (Data Archiving and Network Services) http://www.dans.knaw.nl Kutatási adatok archiválása a saját fejlesztésű EASY
DANS és Narcis Burmeister Erzsébet HUNOR találkozó, Budapest 2013. március 13. DANS DANS (Data Archiving and Network Services) http://www.dans.knaw.nl Kutatási adatok archiválása a saját fejlesztésű EASY
Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális
 Ipari Ökológia pp. 17 22. (2015) 3. évfolyam, 1. szám Magyar Ipari Ökológiai Társaság MIPOET 2015 Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális elegyekre* Tóth András
Ipari Ökológia pp. 17 22. (2015) 3. évfolyam, 1. szám Magyar Ipari Ökológiai Társaság MIPOET 2015 Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális elegyekre* Tóth András
Képrekonstrukció 2. előadás
 Képrekonstrukció 2. előadás Balázs Péter Képfeldolgozás és Számítógépes Grafika tanszék Szegedi Tudományegyetem Az atomszerkezet Atommag (nukleusz): {protonok (poz. töltés) és neutronok} = nukleonok Keringő
Képrekonstrukció 2. előadás Balázs Péter Képfeldolgozás és Számítógépes Grafika tanszék Szegedi Tudományegyetem Az atomszerkezet Atommag (nukleusz): {protonok (poz. töltés) és neutronok} = nukleonok Keringő
SUPPLEMENTAL DATA. Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli.
 SUPPLEMENTAL DATA Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli. B. Douzi, Y.R. Brunet, S. Spinelli, V. Lensi, P. Legrand, S. Blangy,
SUPPLEMENTAL DATA Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli. B. Douzi, Y.R. Brunet, S. Spinelli, V. Lensi, P. Legrand, S. Blangy,
SOLiD Technology. library preparation & Sequencing Chemistry (sequencing by ligation!) Imaging and analysis. Application specific sample preparation
 SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
PIACI HIRDETMÉNY / MARKET NOTICE
 PIACI HIRDETMÉNY / MARKET NOTICE HUPX DAM Másnapi Aukció / HUPX DAM Day-Ahead Auction Iktatási szám / Notice #: Dátum / Of: 18/11/2014 HUPX-MN-DAM-2014-0023 Tárgy / Subject: Változások a HUPX másnapi piac
PIACI HIRDETMÉNY / MARKET NOTICE HUPX DAM Másnapi Aukció / HUPX DAM Day-Ahead Auction Iktatási szám / Notice #: Dátum / Of: 18/11/2014 HUPX-MN-DAM-2014-0023 Tárgy / Subject: Változások a HUPX másnapi piac
Gottsegen National Institute of Cardiology. Prof. A. JÁNOSI
 Myocardial Infarction Registry Pilot Study Hungarian Myocardial Infarction Register Gottsegen National Institute of Cardiology Prof. A. JÁNOSI A https://ir.kardio.hu A Web based study with quality assurance
Myocardial Infarction Registry Pilot Study Hungarian Myocardial Infarction Register Gottsegen National Institute of Cardiology Prof. A. JÁNOSI A https://ir.kardio.hu A Web based study with quality assurance
Performance Modeling of Intelligent Car Parking Systems
 Performance Modeling of Intelligent Car Parking Systems Károly Farkas Gábor Horváth András Mészáros Miklós Telek Technical University of Budapest, Hungary EPEW 2014, Florence, Italy Outline Intelligent
Performance Modeling of Intelligent Car Parking Systems Károly Farkas Gábor Horváth András Mészáros Miklós Telek Technical University of Budapest, Hungary EPEW 2014, Florence, Italy Outline Intelligent
Nemzetközi vállalat - a vállalati szoftvermegoldások egyik vezető szállítója
 Nemzetközi vállalat - a vállalati szoftvermegoldások egyik vezető szállítója A Novell világszerte vezető szerepet tölt be a Linux-alapú és nyílt forráskódú vállalati operációs rendszerek, valamit a vegyes
Nemzetközi vállalat - a vállalati szoftvermegoldások egyik vezető szállítója A Novell világszerte vezető szerepet tölt be a Linux-alapú és nyílt forráskódú vállalati operációs rendszerek, valamit a vegyes
Certificate no./bizonyítvány száma: ÉlfF/200-29/2017. ÁLLATEGÉSZSÉGÜGYI BIZONYÍTVÁNY
 Certificate no./bizonyítvány száma: ÉlfF/20029/2017. ÁLLATEGÉSZSÉGÜGYI BIZONYÍTVÁNY A. B. Certificate no./bizonyítvány száma: ÉlfF/20029/2017. C. D. Certificate no./bizonyítvány száma: ÉlfF/20029/2017.
Certificate no./bizonyítvány száma: ÉlfF/20029/2017. ÁLLATEGÉSZSÉGÜGYI BIZONYÍTVÁNY A. B. Certificate no./bizonyítvány száma: ÉlfF/20029/2017. C. D. Certificate no./bizonyítvány száma: ÉlfF/20029/2017.
Közlekedésbiztonsági Szakmai Nap
 Közlekedésbiztonsági Szakmai Nap 2011. November 23. 1 Svinger László üzletágvezető 3M. Minnesota Mining & Manufacturing 2 3M technológiák Ab Abrasives Ac Acoustics Ad Adhesives Am Advanced Materials An
Közlekedésbiztonsági Szakmai Nap 2011. November 23. 1 Svinger László üzletágvezető 3M. Minnesota Mining & Manufacturing 2 3M technológiák Ab Abrasives Ac Acoustics Ad Adhesives Am Advanced Materials An
Adatkezelő szoftver. Továbbfejlesztett termékvizsgálat-felügyelet Fokozott minőség és gyártási hatékonyság
 Adatkezelő szoftver ProdX Inspect szoftver Fokozott termelékenység Páratlan termékminőség Magas fokú biztonság Teljesen átlátható folyamatok Továbbfejlesztett termékvizsgálat-felügyelet Fokozott minőség
Adatkezelő szoftver ProdX Inspect szoftver Fokozott termelékenység Páratlan termékminőség Magas fokú biztonság Teljesen átlátható folyamatok Továbbfejlesztett termékvizsgálat-felügyelet Fokozott minőség
Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley M. Lemon, Lawrence M. Pfeffer, Kui Li
 Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
FÖLDRAJZ ANGOL NYELVEN
 Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
Számítógéppel irányított rendszerek elmélete. Gyakorlat - Mintavételezés, DT-LTI rendszermodellek
 Számítógéppel irányított rendszerek elmélete Gyakorlat - Mintavételezés, DT-LTI rendszermodellek Hangos Katalin Villamosmérnöki és Információs Rendszerek Tanszék e-mail: hangos.katalin@virt.uni-pannon.hu
Számítógéppel irányított rendszerek elmélete Gyakorlat - Mintavételezés, DT-LTI rendszermodellek Hangos Katalin Villamosmérnöki és Információs Rendszerek Tanszék e-mail: hangos.katalin@virt.uni-pannon.hu
Baldwin St S Ashburn Rd Baldwin St N Anderson St Thickson Rd N Exhibit 'A' to Amendment to the Whitby Official Plan Exhibit 1 Deferral #2 Modification
 Baldwin St S Ashburn Rd Baldwin St N Anderson St Thickson Rd N Exhibit 'A' to Amendment to the Whitby Official Plan Exhibit 1 Modification #3 Brawley Rd W Brawley Rd E U Add: Community Central Area Add:
Baldwin St S Ashburn Rd Baldwin St N Anderson St Thickson Rd N Exhibit 'A' to Amendment to the Whitby Official Plan Exhibit 1 Modification #3 Brawley Rd W Brawley Rd E U Add: Community Central Area Add:
FÜGGETLEN KONTROLLOK ALKALMAZÁSA A NAPI GYAKORLATBAN
 FÜGGETLEN KONTROLLOK ALKALMAZÁSA A NAPI GYAKORLATBAN Debreczeni Lóránd, Kovácsay Anna, Komlovszki Éva Fővárosi Önkormányzat Szent Imre Kórház, Központi Laboratórium Bio-Rad, Budapest 2006 Establishing
FÜGGETLEN KONTROLLOK ALKALMAZÁSA A NAPI GYAKORLATBAN Debreczeni Lóránd, Kovácsay Anna, Komlovszki Éva Fővárosi Önkormányzat Szent Imre Kórház, Központi Laboratórium Bio-Rad, Budapest 2006 Establishing
MEDINPROT Gépidő Pályázat támogatásával elért eredmények
 A kisszögű röntgenszórási módszer fejlesztése fehérjék oldatfázisú mérésére Bóta Attila, Wacha András, Varga Zoltán MTA TTK Biológiai Nanokémia Kutatócsoport 1117 Bp. Magyar Tudósok krt. 2. MEDINPROT Gépidő
A kisszögű röntgenszórási módszer fejlesztése fehérjék oldatfázisú mérésére Bóta Attila, Wacha András, Varga Zoltán MTA TTK Biológiai Nanokémia Kutatócsoport 1117 Bp. Magyar Tudósok krt. 2. MEDINPROT Gépidő
A jövedelem alakulásának vizsgálata az észak-alföldi régióban az 1997-99. évi adatok alapján
 A jövedelem alakulásának vizsgálata az észak-alföldi régióban az 1997-99. évi adatok alapján Rózsa Attila Debreceni Egyetem Agrártudományi Centrum, Agrárgazdasági és Vidékfejlesztési Intézet, Számviteli
A jövedelem alakulásának vizsgálata az észak-alföldi régióban az 1997-99. évi adatok alapján Rózsa Attila Debreceni Egyetem Agrártudományi Centrum, Agrárgazdasági és Vidékfejlesztési Intézet, Számviteli
Meteorológiai ensemble elırejelzések hidrológiai célú alkalmazásai
 Meteorológiai ensemble elırejelzések hidrológiai célú alkalmazásai Országos Vízjelzı Szolgálat CSÍK András Országos Vízjelzı Szolgálat Budapest, 214. február 27. Ensemble elırejelzések elınye Determinisztikus
Meteorológiai ensemble elırejelzések hidrológiai célú alkalmazásai Országos Vízjelzı Szolgálat CSÍK András Országos Vízjelzı Szolgálat Budapest, 214. február 27. Ensemble elırejelzések elınye Determinisztikus
Az Ipoly árvízi előrejelző rendszer
 Az Ipoly árvízi előrejelző rendszer Papanek László Katonáné Kozák Edit Pálfi Gergely Magyar Hidrológiai Társaság XXXIII. Vándorgyűlés 2015.06.02. A feladat közös, integrált, valós idejű hidrológiai előrejelző
Az Ipoly árvízi előrejelző rendszer Papanek László Katonáné Kozák Edit Pálfi Gergely Magyar Hidrológiai Társaság XXXIII. Vándorgyűlés 2015.06.02. A feladat közös, integrált, valós idejű hidrológiai előrejelző
PARK ATRIUM IRODAHÁZ PARK ATRIUM OFFICE BUILDING
 01 0.floor/0.emelet 1550 32 2969 1 1 +600 8 91 1650 +600 65 95 low 44 high 40-1 35-20 18,4 5 131,64 m 2 1 5 1 5 33, m 2 15,8 m 2 32,3 m 2 PARK ATRIUM IROAHÁZ PARK ATRIUM BUILING 15,00 m 2 see service block
01 0.floor/0.emelet 1550 32 2969 1 1 +600 8 91 1650 +600 65 95 low 44 high 40-1 35-20 18,4 5 131,64 m 2 1 5 1 5 33, m 2 15,8 m 2 32,3 m 2 PARK ATRIUM IROAHÁZ PARK ATRIUM BUILING 15,00 m 2 see service block
