SUPPLEMENTARY INFORMATION
|
|
|
- Gergely Pintér
- 6 évvel ezelőtt
- Látták:
Átírás
1 doi: /nature14095 Supplementary Table 1: MAGE oligonucleotides. Name Sequence Strategy acpp.high.1.ny t*g*cttaacgcccagctgttcgccgataattttcttaacgcgttcctagatagtgctcatactcttaaatttcctatcaaaactcgctttc Tolerant acpp.low.1.ny a*t*tttctcagcttcttcgtccggaatctcagtatcaaactcttcctacagagccattaccagctcaacggtgtcaagagaatccgcgccc Tolerant acpp.med.1.ny g*t*aacttcttcctgcttaacgcccagctgttcgccgataattttctaaacgcgttcttcgatagtgctcatactcttaaatttcctatca Tolerant asps.high.1.ny a*g*gtagcggtatttcagacgcgcttcttcggtgttgacgtggttctagtcaagcggcagaacatctgcgcggttgatgatagtcagcgag Tolerant asps.low.1.ny g*c*aaagctcggtgcttcagtcatcagacacgctgccgccgtggtctacgggaaggcgataacgtcacggatattgtcggtgccggtcagc Tolerant asps.med.1.ny t*a*tttcagacgcgcttcttcggtgttgacgtggttagagtcaagctacagaacatctgcgcggttgatgatagtcagcgaggacgccagc Tolerant can.high.1.ny a*t*agctcattgctcggcgaaatgccgcaagagcgccgtctggattagttgtgtgaactgaacgtcatggaacaggtgtataacctgggcc Tolerant can.low.1.ny g*t*taccgtcacgggatttccaacctcaagctgaaacacgccaactagaaataaaaatgccatgccggatgcaacacatccggcaacttca Tolerant can.med.1.ny a*a*ctggggcttatcaacaactggctgctgcatatccgcgatatctagttcaaacatagctcattgctcggcgaaatgccgcaagagcgcc Tolerant cyss.high.1.ny g*g*ttcttgctccagcaggcccaatacagcggaaagtttacgcagctaagatgccattgcattcgccgctgccatatcttctgctttcaga Tolerant cyss.low.1.ny c*a*atagcgcaattacttacgacgccaggtggtcccttgcgggccctattccagcacgatccccatctcgttaagacgatcacgcgccgca Tolerant cyss.med.1.ny c*c*tttggcgatcagttgttcagtgagttcaataatttctgcgatctagtgcgtcgcgcgcggctccatatccgggcgcagaatgttcaaa Tolerant eno.high.1.ny g*t*gaattcttcagaggtgaacgctttgttgccttcgccagccagctagtatttaccatctttgtagaattcagaagctgcgcagtccatc Tolerant eno.low.1.ny t*t*acgaccgttgtacggtgctttttcgcccagagcttcttcgatctaaatcagctggttgtatttagcaacacggtcagaacggctcata Tolerant eno.med.1.ny a*g*gaaacgggatttgtcgccatcgcgcagttccagagcttcacgctaaccagtagaagcacctgacggagcagctgccataccgacgaaa Tolerant era.high.1.ny t*c*ttccgggaagtgatgagtcgcttcaggtagatgcttacgcacctatgccgcaatagtgtcaacattcagcccggtttcggcagagatt Tolerant era.low.1.ny c*a*tcggagttactcttaaagatcgtcaacgtaaccgagactgcgctatgcgcgttcgtcgtcggcccaaccggatttcacttttacccac Tolerant era.med.1.ny c*a*gcgggtgccttcaacgacaaaaatcaccagctcaacatcgccctaagagctgctcgccgctttgttcatcaggcggttaatggcgcgt Tolerant fabg.high.1.ny a*t*accggcattattgaccaggatatccacttcaccaaattctgcctaaattttttccagaacagattcgatagatgccgggtcggtcaca Tolerant fabg.low.1.ny a*c*atgagcattttgtgcaaatcgcggtcagaccatgtacatcccctagttcacatgcaaagtttcacccgtgatgtaagctgcttcgtcg Tolerant fabg.med.1.ny t*c*tttcattcgcattaacaggttatcacgagtgataccggcattctagaccaggatatccacttcaccaaattctgcgcgaattttttcc Tolerant glns.high.1.ny g*g*gttaatcaccgacaggaaatcatccgcagcacctgggttaggctagctgaacagacggtcatacaaacggatttcaaccggcagcgca Tolerant glns.low.1.ny a*c*cggtttttccgccgtagaatggcggctatcgaggcagaagtactattcacgctcaaactggaatgctttacccgcaaccgcatctttc Tolerant glns.med.1.ny t*c*gcggtacgggctgtttttacccggttgcgtcagggtgccgcgctattcgcggatctgttccggcgtcagttcatcaacgtacgccagg Tolerant gros.high.1.ny c*c*atcgttgaaaataacgatgtcgccaactttcacatccagcggctacacttcgccattttcaaggatacggccattgccgacagccagc Tolerant gros.low.1.ny a*c*atccagcggcttcacttcgccattttcaaggatacggccattctagacagccagcacttcgccgcgggtggatttagccgctgcagag Tolerant gros.med.1.ny a*c*gatgccgccagcagatttagtttcaacttctttacgcttgacctacacgcgatcatgcaatggacgaatattcattgataactctcct Tolerant isph.high.1.ny c*g*gttatgtaccacttcgtgacggacatatatcggtgcgccgtactaggccagcgcgttttcaacaatgctgatagcgcggtctaccccg Tolerant isph.low.1.ny c*c*cagctgctgcaaacgtgccaccacattctgcaccagaatatcctaagccgatgcgcccgcagtcacgccgacgcatttaacctctttc Tolerant isph.med.1.ny g*t*caaatcgcgactttttgcttcgttacgtaccgcctgagaaacctagtgtgcggagaaaatcaggatcgcgccgtccggtacttcgcta Tolerant lepb.high.1.ny t*t*tttcaacgttgctttatccagtgagtccccggcagccgcctgctacgctgcctgacgttcccgccgtttaggtgcgaaaaagaattta Tolerant lepb.low.1.ny c*a*taacggcgacaacgtgaacgaagatggctattaatggatgccctaaatgcgacttaagcgcagaccagtcggccattcgccttcttgc Tolerant lepb.med.1.ny g*c*gttttcacacgcctggccggaactgcatcccggttgaatcgtctactcttttgagaccggatcgtaagtgactttatcgcccggtaaa Tolerant murd.high.1.ny a*t*accgggactggcgacaatcagatctgccgccatcagccattcctaattcagactgcccgtgtggcgttctacggcttcgggtaattta Tolerant murd.low.1.ny t*a*aacgcatcaacctaactccttcgccagacgggcaaactcattctatcgttgttcaaagttcttgaactgatcaaggctggcacaggct Tolerant murd.med.1.ny g*a*ggtggtttccagctggaagctcgacagttccagcacgtacagctaacactcatcatccagtagcatcaacgcaggcaggccaatattg Tolerant rplj.high.1.ny t*t*cgcgaactctttgaacagacgagcagcagcgcccgggtgttcctaagagtatgcaatcagggtcggaccaacaaacgcgtctttcagg Tolerant rplj.high.2.ny g*c*cagagtacgaaccagtttgccagccgaagcttctttcatggtctacatcaggcgtgcaattgcttcttcgtaggtcggcagagttgcc Tolerant rplj.low.1.ny g*c*agcagcgcccgggtgttccatagagtatgcaatcagggtcggctaaacaaacgcgtctttcaggcactcgaacggagtaccttcaaca Tolerant rplj.low.2.ny t*c*tttcatggttgccatcaggcgtgcaattgcttcttcgtaggtctacagagttgccaggcggtcgatctgagacgccgggatcagctca Tolerant rplj.med.1.ny g*t*cattttatctacagttacgccacgggaatccgcaactactgcctacagcgcgcctttggctacttcgctgacttcagcaacaatcgct Tolerant rplj.med.2.ny t*c*agtcattttatctacagttacgccacgggaatccgcaactacctaagacagcgcgcctttggctacttcgctgacttcagcaacaatc Tolerant rplm.high.1.ny t*t*tggcaacatgcctttaaccgcgatttcaatcacacgctcaggctagcgagcaatcatctcttcaaaggtcgcttgtttgataccaccg Tolerant rplm.low.1.ny g*c*cattgcctataatcccgattagatgtcaagaacttgcggttgctatgccgcgtggttgtgctcgttacccgcgtaaactttcagttta Tolerant rplm.med.1.ny a*g*ttcagtagccagacggcccagagttttaccggtcgcgtcaacctaataccagtcgcgttttacggtttctggtttagctgtaaaagtt Tolerant rpls.high.1.ny a*c*taccggagagtgagtctggaagacacgctcaacgccttcgccctaggaaattttacgaacagtgaatgcagagtgcagaccgcggtta Tolerant rpls.low.1.ny t*a*gttaagacgctctttgatacgagcagccttaccagtacgctcctacaggtagtacagtttagctttacgaacagcaccacgacgtttg Tolerant rpls.med.1.ny g*t*atcacccggacggaaggaaggtacgtcctgcttcatctgctcctattcaagttgcttaataatgttgctcataatttaatctcttatc Tolerant rpmd.high.1.ny c*c*caggccaagcagcgttgccttgtgtttcggcagacgaccgatctaactgcgggtttgagtaattttaatagtctttgccatggtttat Tolerant rpmd.low.1.ny g*c*aggagtatcctcgcgctctacggtgtgaccaatacgacgcagctacaggccaagcagcgttgccttgtgtttcggcagacgaccgatt Tolerant rpmd.med.1.ny g*c*cttgtgtttcggcagacgaccgattgcactgcgggtttgagtctatttaatagtctttgccatggtttatttccccagaatttcttca Tolerant rpoa.high.1.ny t*a*ggcaatacgctccacagggctgtagcatgcgtcgaccagcagctagccgattgggcgctcatcttcttccgaatgaattcgggtagaa Tolerant rpoa.low.1.ny g*g*ccagttttccaggcgcatgcccagagacagtccacgggaagcctacacgtctttaatctcagtaagagattttttaccaaggttaggc Tolerant rpoa.med.1.ny a*g*gttgagcaggatttccaggatatcttcctgaacgccttctttctagctgtactcatgtagtacaccatcaatctcaacctcggtcacc Tolerant rpsb.high.1.ny a*a*ctattttgggggagttatcaagccttattactcagcttctacctagctttcttccgcctgggaagccagatcctgagaacggccttca Tolerant rpsb.low.1.ny g*c*agcaacagcgcccaggtacagggtcacagcacggattgcgtcctagttacccgggataacgaagtcaacaccgtccggatcagagttg Tolerant rpsb.med.1.ny t*t*gttcagttcagccagagcttcgttgaacatcggtacagttttctaaaggttgatgatgtgaactttgttacgcgcaccgaagatgaac Tolerant rpsd.high.1.ny a*a*cgtaccttccatcttgccagcatcaacttccagccaggttggctattcacgctgctcagccagctccagagcggctttcacgcgagac Tolerant rpsd.low.1.ny g*c*attgtgtcctctctttggtactaagctttacttggagtaaagctagacgatcaggtgttcgttaatgtccgcagacagatcagaacgc Tolerant rpsd.med.1.ny c*g*aactttttgcttttcacgcaactgcacaccatagtcagacagctacggtttacgcgcaccgtgctggccaggagcttgttcaatttta Tolerant 1
2 thrs.high.1.ny t*c*ctccaattgtttaagactgcggctgcgaatctcttgttgcagctactcgatcacttcatttacgtccatgcttcccaggtctttacca Tolerant thrs.low.1.ny t*c*tttaccacggcgggtgcgaacggcaactttgcctgattccacctatttatcaccacagaccagcatatatgggacgcgacgcaaagtg Tolerant thrs.med.1.ny t*g*ccagctgactttcttcttaatgacgtcgtagtttttctcagcctactcatgcatccgcttctcgagtgcttcgacatcttcctgggtt Tolerant trmd.high.1.ny t*t*tgcgccttcacccgccgcggcttttgctgcatgaatggcgtcctacaagggttgcaccatcattaacatccccggtccgccgccgtaa Tolerant trmd.high.2.ny g*g*tccgccgccgtaaggacgatcgtccacggtacggtgccggtcctacgtgaagtcgcgaggactccagctctggatgctcagcaggcca Tolerant trmd.low.1.ny a*t*ctcttatcctgggtaaactgatatctcgggggcttacgccatctaatcatgtttatgttgctgttgtgcgtgttccgttttgaactcc Tolerant trmd.low.2.ny a*a*tttaatctcttatcctgggtaaactgatatctcgggggcttactacatcccatcatgtttatgttgctgttgtgcgtgttccgttttg Tolerant trmd.med.1.ny c*c*cgccgcggcttttgctgcatgaatggcgtcccgcaagggttgctacatcattaacatccccggtccgccgccgtaaggacgatcgtcc Tolerant trmd.med.2.ny t*c*acccgccgcggcttttgctgcatgaatggcgtcccgcaagggctacaccatcattaacatccccggtccgccgccgtaaggacgatcg Tolerant acpp.low.3 a*a*cgaccgcctggagatgttcacttacgcctggtggccgttgatctaatcaatggcagcctgaacggtggtgattttctcagcttcttcg Tolerant can.high.20.a c*a*gtggatgtactcgaagttgaacacattattatctgtggccactagggttgcggcggcgtacaagccgcagttgaaaacccggaactgg Tolerant can.high.20.b g*c*cgtctggataccttgtgtgaactgaacgtcatggaacaggtgtagaacctgggccactccaccattatgcaatcagcgtggaaacgcg Tolerant can.low.2 a*c*ctggtcattcacactgacctgaactgcctttccgtggttcagtaggcagtggatgtactcgaagttgaacacattattatctgtggcc Tolerant dnax.high.20 a*c*ttcgtcgatcagataaactttgaaacgaccacgcgccggagcctactggacgttatccagcaggtcgcgggtatcttcaactttggtg Tolerant dnax.low.5 g*c*gttaaagctgtggcgcgacagcatatgcacttcgtcgatcagctaaactttgaaacgaccacgcgccggagcgtactggacgttatcc Tolerant dnax.med.16 g*g*cgcatacggtaattctttgcgaccaatcaacagcgtctgatactaaagctgaatatccgtcggcggtatggtgcgcgccagttcacgc Tolerant dxs.high.20 t*g*ccgaaagagagtttaccgaaactctgcgacgaactgcgccgctagttactcgacagcgtgagccgttccagcgggcacttcgcctccg Tolerant dxs.low.6 t*c*gcctccgggctgggcacggtcgaactgaccgtggcgctgcactaggtctacaacaccccgtttgaccaattgatttgggatgtggggc Tolerant dxs.med.11 c*a*ataagtattaataggcccctgatgagttttgatattgccaaatagccgaccctggcactggtcgactccacccaggagttacgactgt Tolerant eno.high.20 t*g*ctcaccaccgttgatgatgttcatcatcggaaccggcatagactatttgcccggagtaccgttcagttcagcgatgtgctcgtacagc Tolerant eno.low.2.a a*t*agagtatttgcccggagtaccgttcagttcagcgatgtgctcctacagcggcatacctttagcagctgcagcagctttggcgttagcc Tolerant eno.low.2.b g*c*gataacagccagagcttcagcgttggaacccaggttcggcgcctagccaccttcgtcaccaacagcagtgttcatgcctttcgctttc Tolerant fabg.high.20 c*t*gcgcgcaacatgagcattttgtgcaaatcgcggtcagaccatctacatcccgccgttcacatgcaaagtttcacccgtgatgtaagct Tolerant fabg.low.1 a*c*catgtacatcccgccgttcacatgcaaagtttcacccgtgatctaagctgcttcgtcggatgccaggaatgcaaccgcgttggcgatt Tolerant fabg.med.18 g*g*gtcggtcacattcaacatcagacctttgccgttggcacctaactaatcactgatcgcctgagcgccattttcactggtcgcagtgcca Tolerant isph.low.1 t*g*ttctgccagggcgcgtaccgcttcctgacggttagtcgtggcctagcagatgtcatctttgcgcggaccgacaattttcgggaagcgt Tolerant isph.med.10 t*c*gtccggcgattcgaccagatacattcccccttccgggttactctactggcccattgtcccttccacttccgggtgcccggcgtgaccg Tolerant isph.med.11 t*t*tttgaccgtcagtttccacacatcgtccggcgattcgaccagctacattcccccttccgggttactgtactggcccattgtcccttcc Tolerant lspa.high.20 a*a*cagcgggaccgtatcccccagagcaaagttctggaggatcagctatttgctgcccagatcgataatcagcacgactaccaccagccac Tolerant lspa.low.3 c*c*gctatcggcaaggaaactaaacgccgcgccatagttacgcgcctaatgcagattaagcgacgggaacagcgggaccgtatcccccaga Tolerant lspa.med.15 c*g*ctgccagccgccgctatcggcaaggaaactaaacgccgcgccctagttacgcgcataatgcagattaagcgacgggaacagcgggacc Tolerant phet.high.20 t*c*atagaaatcaacggtctcttttgccaggttccagtgctcttcctaacggttaccgcaaatcacaccggctaacatcagatcctgacga Tolerant phet.low.2 t*t*accggtcaggtcgagaacggattcaagatcgcctttcaaatcctagaaatcaacggtctcttttgccaggttccagtgctcttcgtaa Tolerant phet.med.14 c*c*gctttcgaaaatgcgcacacggttctgctgacggttctggttctacaccacggttgccagcaggccagtccacagagaaagacgcatt Tolerant rplb.high.20 a*g*acgcagggtgacataagcaccatcacgagcaacgatctgaacctaagtaccagcggaacgtgccagctgaccgcctttacctggtttc Tolerant rplb.low.1.a g*c*ttctactttacgcatttcaccagaacgcagacgcagggtgacctaagcaccatcacgagcaacgatctgaacgtaagtaccagcggaa Tolerant rplb.low.1.b g*a*ctgaatctggtcgccagctttcaggcctttaggggccaggatctaacggcgttcaccgtctttgtacagaaccagcgcgatgttcgcg Tolerant rplj.low.4 a*a*ctctttgaacagacgagcagcagcgcccgggtgttccatagactatgcaatcagggtcggaccaacaaacgcgtctttcaggcactcg Tolerant rplj.med.10 c*c*ttcaacagcacggcgcagcagggtgttacgaacaacacgcatctatacgccagcttcgcgacctgctttacgcagttcagtcatttta Tolerant rplj.med.13 g*a*agcttctttcatggttgccatcaggcgtgcaattgcttcttcctaggtcggcagagttgccaggcggtcgatctgagacgccgggatc Tolerant rplm.low.2 g*c*gttcagaacgatgatgtaatcaccggtatctacgtgcggagtctattccgctttgtgcttaccgcgcaggcgacgagccagttcagta Tolerant rplm.low.4 t*t*gttgccggttacagcaactttgtcagcgttcagaacgatgatctaatcaccggtatctacgtgcggagtgtattccgctttgtgctta Tolerant rplm.low.5 t*c*agtagccagacggcccagagttttaccggtcgcgtcaacaacctaccagtcgcgttttacggtttctggtttagctgtaaaagttttc Tolerant rpls.low.1 a*g*acgctctttgatacgagcagccttaccagtacgctcacgcagctagtacagtttagctttacgaacagcaccacgacgtttgacagaa Tolerant rpls.low.2 c*g*ctctttgatacgagcagccttaccagtacgctcacgcaggtactacagtttagctttacgaacagcaccacgacgtttgacagaaatg Tolerant rpmb.low.4 c*t*cacgaatacctttagccatgatttatttcctctaagtacttactacttttcgccacgggcacgcagttcagccagaactgtatcgatg Tolerant rpsd.low.1 a*a*gtcggtgccctcacgacggctcagcttgagcttaggacccaactatcttgccattttctttctccaacaaacctggaaaacgaggcgt Tolerant rpsd.low.5 t*t*cgctttctcacgaatgcttacaacgtcattcggactaacctgctaagaagcgatgttaacaacacgaccgtttaccataattgcttta Tolerant rpsk.low.8 c*c*tggacccggacctttaaccataacttccagattcttgatgccctattctttcacggcgtcagcgcaacgctctgctgcaacctgagct Tolerant rpsl.low.1 a*g*gatcacggagtgctcctgcaggttgtgaccttcaccaccgatctaggaagtcacttcgaaaccgttagtcagacgaacacggcatact Tolerant rpsl.low.2 c*a*tactttacgcagcgcggagttcggttttttaggagtggtagtctatacacgagtacatacgccacgtttttgcgggcatgcttccagc Tolerant rpsp.low.2 a*a*gcgaccgttgcgtgcattacggctgtcagcgacaacaacctgctagaacggacgctttttagcgccgtgacgtgctaaacgaatagtt Tolerant secy.high.20 t*c*cataatcacgacaacaacgataagcagtgaggtcccaccgaactagaacggtactttcattgcatcacgcatgaactccgggatcagg Tolerant secy.low.1 g*t*agcaataccgatcgactggaatattgccagcaccagagtaccctagcgggtgtactggctgatcttacgacgaccagactccccttct Tolerant secy.med.18 g*c*agacgcatagagtaacacataaagcggttgcccaggctgcaactacagcgaaattgttgtcagccagttccaaccagtaccgcccccg Tolerant topa.high.20 t*c*ggccagataacgcagtttttccggcagacggtcgcggaagcgctaaagctcttccaccagtggcgcacgcgtttcacgcgatttcggg Tolerant topa.low.5 t*t*ggcagccaggaacacaccggcagcaccgtcacgcagcacgaactaagcatctgatttttcgcacggcagctcaggtaatggcaccgga Tolerant topa.med.14 a*a*aatcttacgtgtgtttttacactcttcgttggtgcaggccatctatttaccgaaacgccccattttcaggtgcatttcagagccacat Tolerant trmd.low.2 t*c*ccgcaagggttgcaccatcattaacatccccggtccgccgccctaaggacgatcgtccacggtacggtgccggtcatgcgtgaagtcg Tolerant trmd.low.4 t*c*gctgacgcccgcttgatcaagcttgcgtccctgtggtgacagctaaatcacctttgcgccttcacccgccgcggcttttgctgcatga Tolerant trmd.med.16 t*g*gatgctcagcaggccatttttaactgcccggccagttaccccctaatcggtaattgcgcggaacatttcaggaaacaggctaattatg Tolerant 2
3 tyrs.low.1 t*a*ctgtttgttcagacaggcgaagtcataaccctgcaacaggttctaggaaaactcagtgaacgaaatcccctgatcttcacggttgaga Tolerant tyrs.med.18 a*t*atcgcgcaggaaggtcagcacattcatattgccgaaccagtcctagttgttcgccgcgatagcagagttttctccacagtcgaaatcg Tolerant ftsl.n.1tag c*g*atcctttaactttgcttagagcttctgtcactctgctgatctacatgcattcgtcctctctgcaatacgcagaactgaactacgggca Amino-terminal ftsl.n.2tag c*g*atcctttaactttgcttagagcttctgtcactctgctgatctactacatgcattcgtcctctctgcaatacgcagaactgaactacgg Amino-terminal ftsl.n.3tag g*a*tcctttaactttgcttagagcttctgtcactctgctgatctactactacatgcattcgtcctctctgcaatacgcagaactgaactac Amino-terminal ftsi.n.1tag t*t*cttcctgacgttttggtttctgcgttttcgccgctgctttctacatgcgtcgcgtttatccttatttttgcactacgatattttcttg Amino-terminal ftsi.n.2tag t*t*cttcctgacgttttggtttctgcgttttcgccgctgctttctactacatgcgtcgcgtttatccttatttttgcactacgatattttc Amino-terminal ftsi.n.3tag c*t*tcctgacgttttggtttctgcgttttcgccgctgctttctactactacatgcgtcgcgtttatccttatttttgcactacgatatttt Amino-terminal murd.n.1tag g*c*ccaggccgataatgacgacatttttaccctgataatcagcctacatgattaacgtaccttcagcgttgccagaccaatcagaaccagc Amino-terminal murd.n.2tag g*c*ccaggccgataatgacgacatttttaccctgataatcagcctactacatgattaacgtaccttcagcgttgccagaccaatcagaacc Amino-terminal murd.n.3tag c*c*aggccgataatgacgacatttttaccctgataatcagcctactactacatgattaacgtaccttcagcgttgccagaccaatcagaac Amino-terminal ftsa.n.1tag a*a*tctccagtcctactaccagttttctgtccgtcgccttgatctacattgttgttctgcctgtgcctgattttgttgctgagtagattcc Amino-terminal ftsa.n.2tag c*t*ccagtcctactaccagttttctgtccgtcgccttgatctactacattgttgttctgcctgtgcctgattttgttgctgagtagattcc Amino-terminal ftsa.n.3tag t*c*cagtcctactaccagttttctgtccgtcgccttgatctactactacattgttgttctgcctgtgcctgattttgttgctgagtagatt Amino-terminal ftsz.n.1tag g*a*ctttaatcaccgcgtcattggtaagttccattggttcaaactacatagtttctctccgatttgtgcctgtcgcctgaggccgtaatca Amino-terminal ftsz.n.2tag g*a*ctttaatcaccgcgtcattggtaagttccattggttcaaactactacatagtttctctccgatttgtgcctgtcgcctgaggccgtaa Amino-terminal ftsz.n.3tag a*c*tttaatcaccgcgtcattggtaagttccattggttcaaactactactacatagtttctctccgatttgtgcctgtcgcctgaggccgt Amino-terminal seca.n.1tag a*t*cgttacgactaccgaaaactttagttaacaatttgattagctacataataaaatctcaaacgccccgcgttgcggagttaataaaatg Amino-terminal seca.n.2tag a*t*cgttacgactaccgaaaactttagttaacaatttgattagctactacataataaaatctcaaacgccccgcgttgcggagttaataaa Amino-terminal seca.n.3tag t*c*gttacgactaccgaaaactttagttaacaatttgattagctactactacataataaaatctcaaacgccccgcgttgcggagttaata Amino-terminal frr.n.1tag t*t*tgtccatgcgtacttcagcatcttttctgatatcgctaatctacacgttacgaatccttgaaaacttgtctcagtcagaccagacagt Amino-terminal frr.n.2tag t*t*tgtccatgcgtacttcagcatcttttctgatatcgctaatctactacacgttacgaatccttgaaaacttgtctcagtcagaccagac Amino-terminal frr.n.3tag t*g*tccatgcgtacttcagcatcttttctgatatcgctaatctactactacacgttacgaatccttgaaaacttgtctcagtcagaccaga Amino-terminal acca.n.1tag c*a*gctctgcaatcggctgttcaaaatcaaggaaattcagactctacatagtattcctgtattagtcaaactccagttccacctgctccga Amino-terminal acca.n.2tag c*a*gctctgcaatcggctgttcaaaatcaaggaaattcagactctactacatagtattcctgtattagtcaaactccagttccacctgctc Amino-terminal acca.n.3tag a*g*ctctgcaatcggctgttcaaaatcaaggaaattcagactctactactacatagtattcctgtattagtcaaactccagttccacctgc Amino-terminal dxs.n.1tag a*t*ccagcgtaataaataaacaataagtattaataggcccctgatgtagagttttgatattgccaaatacccgaccctggcactggtcgac Amino-terminal dxs.n.2tag c*a*gcgtaataaataaacaataagtattaataggcccctgatgtagtagagttttgatattgccaaatacccgaccctggcactggtcgac Amino-terminal dxs.n.3tag a*g*cgtaataaataaacaataagtattaataggcccctgatgtagtagtagagttttgatattgccaaatacccgaccctggcactggtcg Amino-terminal glns.n.1tag g*a*tctgacggataaagttagtcgggcgggcttctgcctcactctacatcgtggattcctcaaagcgtaaacaacgtataacggcgtatga Amino-terminal glns.n.2tag g*a*tctgacggataaagttagtcgggcgggcttctgcctcactctactacatcgtggattcctcaaagcgtaaacaacgtataacggcgta Amino-terminal glns.n.3tag t*c*tgacggataaagttagtcgggcgggcttctgcctcactctactactacatcgtggattcctcaaagcgtaaacaacgtataacggcgt Amino-terminal prsa.n.1tag c*a*gattctttccaccaatggacgcatgcctgaggttcttctcgtgtagcctgatatgaagctttttgctggtaacgccaccccggaacta Amino-terminal prsa.n.2tag a*t*tctttccaccaatggacgcatgcctgaggttcttctcgtgtagtagcctgatatgaagctttttgctggtaacgccaccccggaacta Amino-terminal prsa.n.3tag t*t*ctttccaccaatggacgcatgcctgaggttcttctcgtgtagtagtagcctgatatgaagctttttgctggtaacgccaccccggaac Amino-terminal nade.n.1tag t*a*gctttaaggaagttttgtcttttctgtctggaggggttcaatgtagacattgcaacaacaaataataaaggcgctgggcgcaaaaccg Amino-terminal nade.n.2tag c*t*ttaaggaagttttgtcttttctgtctggaggggttcaatgtagtagacattgcaacaacaaataataaaggcgctgggcgcaaaaccg Amino-terminal nade.n.3tag t*t*taaggaagttttgtcttttctgtctggaggggttcaatgtagtagtagacattgcaacaacaaataataaaggcgctgggcgcaaaac Amino-terminal pgsa.n.1tag g*a*tgacacggaacagtgtaagcaacgtagggatattaaattgctacataatgacgggtaactatctgttgtcagtaagattacccctatg Amino-terminal pgsa.n.2tag g*a*tgacacggaacagtgtaagcaacgtagggatattaaattgctactacataatgacgggtaactatctgttgtcagtaagattacccct Amino-terminal pgsa.n.3tag t*g*acacggaacagtgtaagcaacgtagggatattaaattgctactactacataatgacgggtaactatctgttgtcagtaagattacccc Amino-terminal nrdb.n.1tag a*a*aatgccttatccggcattaaactcccaacaggacacactcatgtaggcatataccaccttttcacagacgaaaaatgatcagctcaaa Amino-terminal nrdb.n.2tag a*t*gccttatccggcattaaactcccaacaggacacactcatgtagtaggcatataccaccttttcacagacgaaaaatgatcagctcaaa Amino-terminal nrdb.n.3tag t*g*ccttatccggcattaaactcccaacaggacacactcatgtagtaggcatataccaccttttcacagacgaaaaatgatcagctca Amino-terminal gltx.n.1tag c*a*gatagcctgttgggcttggcgcgaagcgagttttgattttctacatgaaatggccttacgtttagaaagatgccgacaaccggcaaat Amino-terminal gltx.n.2tag c*a*gatagcctgttgggcttggcgcgaagcgagttttgattttctactacatgaaatggccttacgtttagaaagatgccgacaaccggca Amino-terminal gltx.n.3tag g*a*tagcctgttgggcttggcgcgaagcgagttttgattttctactactacatgaaatggccttacgtttagaaagatgccgacaaccggc Amino-terminal lepb.n.1tag c*a*ccagtgtggcaatcaccagaatcagggcaaacatattcgcctacatgccaactcctaagggttatttgttgtctttgccgacgtgcag Amino-terminal lepb.n.2tag c*a*ccagtgtggcaatcaccagaatcagggcaaacatattcgcctactacatgccaactcctaagggttatttgttgtctttgccgacgtg Amino-terminal lepb.n.3tag c*c*agtgtggcaatcaccagaatcagggcaaacatattcgcctactactacatgccaactcctaagggttatttgttgtctttgccgacgt Amino-terminal trmd.n.1tag t*g*cgcggaacatttcaggaaacaggctaattatgccaatccactacatagcgccgtcttttaccgtttatccggtggtttaaaaaccagg Amino-terminal trmd.n.2tag t*g*cgcggaacatttcaggaaacaggctaattatgccaatccactactacatagcgccgtcttttaccgtttatccggtggtttaaaaacc Amino-terminal trmd.n.3tag c*g*cggaacatttcaggaaacaggctaattatgccaatccactactactacatagcgccgtcttttaccgtttatccggtggtttaaaaac Amino-terminal grpe.n.1tag t*t*ccggggcttgcccctcaggcgttttctgttctttactactctacatgaatttctccgcgtttttttcgcattcatctcgctaacttcg Amino-terminal grpe.n.2tag t*t*ccggggcttgcccctcaggcgttttctgttctttactactctactacatgaatttctccgcgtttttttcgcattcatctcgctaact Amino-terminal grpe.n.3tag c*c*ggggcttgcccctcaggcgttttctgttctttactactctactactacatgaatttctccgcgtttttttcgcattcatctcgctaac Amino-terminal metk.n.1tag a*a*cagtttgagctaaccaaattctctttaggtgatattaaatatgtaggcaaaacacctttttacgtccgagtccgtctctgaagggcat Amino-terminal metk.n.2tag a*g*tttgagctaaccaaattctctttaggtgatattaaatatgtagtaggcaaaacacctttttacgtccgagtccgtctctgaagggcat Amino-terminal metk.n.3tag g*t*ttgagctaaccaaattctctttaggtgatattaaatatgtagtagtaggcaaaacacctttttacgtccgagtccgtctctgaagggc Amino-terminal infb.n.1tag g*g*tctgtcgctctgcggccagcgttttaatcgttacatctgtctacatgctgttccttcctgctacagtttattacgcttcgtcaccgaa Amino-terminal infb.n.2tag g*g*tctgtcgctctgcggccagcgttttaatcgttacatctgtctactacatgctgttccttcctgctacagtttattacgcttcgtcacc Amino-terminal 3
4 infb.n.3tag t*c*tgtcgctctgcggccagcgttttaatcgttacatctgtctactactacatgctgttccttcctgctacagtttattacgcttcgtcac Amino-terminal fmt.n.1tag a*c*tggatcgtctgaaagcccgggcttaaggataagaactaacgtgtagtcagaatcactacgtattatttttgcgggtacacctgacttt Amino-terminal fmt.n.2tag g*g*atcgtctgaaagcccgggcttaaggataagaactaacgtgtagtagtcagaatcactacgtattatttttgcgggtacacctgacttt Amino-terminal fmt.n.3tag g*a*tcgtctgaaagcccgggcttaaggataagaactaacgtgtagtagtagtcagaatcactacgtattatttttgcgggtacacctgact Amino-terminal secy.n.1tag g*c*cacctttggcactttgaaaatctaatcccggttgtttagcctacatctgctacttattcctcgattttaccgccagcagcttcgatag Amino-terminal secy.n.2tag g*c*cacctttggcactttgaaaatctaatcccggttgtttagcctactacatctgctacttattcctcgattttaccgccagcagcttcga Amino-terminal secy.n.3tag c*a*cctttggcactttgaaaatctaatcccggttgtttagcctactactacatctgctacttattcctcgattttaccgccagcagcttcg Amino-terminal rplo.n.1tag c*g*cctttttggagccttcggccggagacagagtatttaaacgctacatctcttactcctcaactttaaccatgaaggaaaccgcgttgat Amino-terminal rplo.n.2tag c*g*cctttttggagccttcggccggagacagagtatttaaacgctactacatctcttactcctcaactttaaccatgaaggaaaccgcgtt Amino-terminal rplo.n.3tag c*c*tttttggagccttcggccggagacagagtatttaaacgctactactacatctcttactcctcaactttaaccatgaaggaaaccgcgt Amino-terminal rpse.n.1tag c*a*gcttttcctgcagttcgccagcttgtttttcgatgtgagcctacatcttacacctctaccttagaactgaaggccagcttcacgggca Amino-terminal rpse.n.2tag c*a*gcttttcctgcagttcgccagcttgtttttcgatgtgagcctactacatcttacacctctaccttagaactgaaggccagcttcacgg Amino-terminal rpse.n.3tag g*c*ttttcctgcagttcgccagcttgtttttcgatgtgagcctactactacatcttacacctctaccttagaactgaaggccagcttcacg Amino-terminal rpsn.n.1tag g*t*tttacttcgcgtgctttcattgattgcttagcctacatttagtaaccctaccttacttgcggaacgggaagtcaaaggcagccagcag Amino-terminal rpsn.n.2tag g*t*tttacttcgcgtgctttcattgattgcttagcctactacatttagtaaccctaccttacttgcggaacgggaagtcaaaggcagccag Amino-terminal rpsn.n.3tag t*t*tacttcgcgtgctttcattgattgcttagcctactactacatttagtaaccctaccttacttgcggaacgggaagtcaaaggcagcca Amino-terminal rpln.n.1tag a*c*cggagttgtcggcgacgttcagcatagtctgttcttggatctacattttagtgctccgctaatgtcaactactactgagacccgaaaa Amino-terminal rpln.n.2tag a*c*cggagttgtcggcgacgttcagcatagtctgttcttggatctactacattttagtgctccgctaatgtcaactactactgagacccga Amino-terminal rpln.n.3tag c*c*ggagttgtcggcgacgttcagcatagtctgttcttggatctactactacattttagtgctccgctaatgtcaactactactgagaccc Amino-terminal rpsq.n.1tag a*a*caacgcgaccttgcagagtacggattttatcggtctacattacgcacccgccttctcgttcagtaaagtcttaacgcgtgcgacatcg Amino-terminal rpsq.n.2tag a*a*caacgcgaccttgcagagtacggattttatcggtctactacattacgcacccgccttctcgttcagtaaagtcttaacgcgtgcgaca Amino-terminal rpsq.n.3tag c*a*acgcgaccttgcagagtacggattttatcggtctactactacattacgcacccgccttctcgttcagtaaagtcttaacgcgtgcgac Amino-terminal rpld.n.1tag g*g*aaacagtcagcgcgctctgcgcgtctttcaatactaattcctacattgctatctccttacgccttcacagctggtttaacgatcaggt Amino-terminal rpld.n.2tag g*g*aaacagtcagcgcgctctgcgcgtctttcaatactaattcctactacattgctatctccttacgccttcacagctggtttaacgatca Amino-terminal rpld.n.3tag a*a*acagtcagcgcgctctgcgcgtctttcaatactaattcctactactacattgctatctccttacgccttcacagctggtttaacgatc Amino-terminal rplc.n.1tag g*a*agatacgggtcatacccacttttttaccgactaaaccaatctacattgtttcaacctctcaatcgctcaatgacctgattaacccagg Amino-terminal rplc.n.2tag g*a*agatacgggtcatacccacttttttaccgactaaaccaatctactacattgtttcaacctctcaatcgctcaatgacctgattaaccc Amino-terminal rplc.n.3tag a*g*atacgggtcatacccacttttttaccgactaaaccaatctactactacattgtttcaacctctcaatcgctcaatgacctgattaacc Amino-terminal fusa.n.1tag g*a*taccgatgttacggtagcgtgcgatgggtgttgtacgagcctacatttgtttcctcgtttatcttttaggcgttcaatttaagtagcc Amino-terminal fusa.n.2tag g*a*taccgatgttacggtagcgtgcgatgggtgttgtacgagcctactacatttgtttcctcgtttatcttttaggcgttcaatttaagta Amino-terminal fusa.n.3tag t*a*ccgatgttacggtagcgtgcgatgggtgttgtacgagcctactactacatttgtttcctcgtttatcttttaggcgttcaatttaagt Amino-terminal rpoc.n.1tag t*t*cggttttagtctgcgctttcagaaactttaataaatctttctacacggatttgctcccgtcggagttagcacaatccgctgccgggtt Amino-terminal rpoc.n.2tag t*t*cggttttagtctgcgctttcagaaactttaataaatctttctactacacggatttgctcccgtcggagttagcacaatccgctgccgg Amino-terminal rpoc.n.3tag c*g*gttttagtctgcgctttcagaaactttaataaatctttctactactacacggatttgctcccgtcggagttagcacaatccgctgccg Amino-terminal ppa.n.1tag t*a*ctcggcacttgtttgccacatatttttaaaggaaacagacatgtagagcttactcaacgtccctgcgggtaaagatctgccggaagac Amino-terminal ppa.n.2tag t*c*ggcacttgtttgccacatatttttaaaggaaacagacatgtagtagagcttactcaacgtccctgcgggtaaagatctgccggaagac Amino-terminal ppa.n.3tag c*g*gcacttgtttgccacatatttttaaaggaaacagacatgtagtagtagagcttactcaacgtccctgcgggtaaagatctgccggaag Amino-terminal dnaa.w6 t*t*ctgtggctggtaactcatcctgcaatcgggcaagacactgctgctaaagcgaaagtgacacggcggactccactcgaacaaaagtcga Functional fusa.f32 a*g*cgccgtcatgaacttcaccgattttatggtttacaccggtgtactacagaatacgttcggtagtagtggttttaccggcgtcgatgtg Functional glyq.y226 c*g*gcagcggattttccagcgccagcagctgctgcgcttctttctcctactgctcgaagcaggtgaacaggaagtccacatccgcgtattc Functional lnt.y388 c*c*gctgtcggcaaatggtattgagcttactgcggctatttgctaggagatcattctcggcgagcaagtgcgcgataacttccgcccggat Functional murg.f243 a*a*cgacgacatccgcccacgcatacgccgccgccatatcatcaatctattccgtcactttatgctgcggttgccccgcttcggcatacgc Functional rpoa.y152 t*g*ggcgctcatcttcttccgaatgaattcgggtagaagccggcacctaaccacgaccgcgctgaactttgatacgcatgctaatagacgc Functional sers.f213 g*t*ctgcttcttcttccagcggacgagtatggaacagatcgccagcctatttcggcagttgacccgtaccgtacagcgtgtcctggttaac Functional thrs.y313 g*c*agtattcacggttctcagaagatgtggtgaacattgcatctttctagttgtcccagtgaccggttttttcccacaggacacggtccat Functional Asterisk (*) signifies a phosphorothioate bond. Oligonucleotides are grouped by strategy (Tolerant, Amino-terminal, and Functional). Table of MAGE oligonucleotides used in this study for the introduction TAG codons within essential genes. Oligonucleotide sequences, location(s) of the encoded TAG codon(s) in the translated product, and design strategies (Tolerant, Amino-terminal, and Functional) are listed. Asterisk (*) signifies a phosphorothioate bond. 4
5 Supplementary Table 2: MAGE pools for the introduction of TAG codons in essential genes. Y NY N C1 C2 F acpp.low.3 acpp.high.1.ny ftsl.n.1tag dnax.high.20 dnax.high.20 dnaa.w6 can.high.20.a acpp.low.1.ny ftsi.n.1tag dnax.low.5 lspa.high.20 fusa.f32 can.high.20.b acpp.med.1.ny murd.n.1tag dnax.med.16 lspa.med.15 glyq.y226 can.low.2 asps.high.1.ny ftsa.n.1tag dxs.high.20 secy.low.1 lnt.y388 dnax.high.20 asps.low.1.ny ftsz.n.1tag dxs.low.6 secy.med.18 murg.f243 dnax.low.5 asps.med.1.ny seca.n.1tag dxs.med.11 topa.med.14 rpoa.y152 dnax.med.16 can.high.1.ny frr.n.1tag eno.high.20 dxs.n.1tag thrs.y313 dxs.high.20 can.low.1.ny acca.n.1tag rplb.high.20 ftsi.n.1tag dxs.low.6 can.med.1.ny dxs.n.1tag rplb.low.1.a murd.n.1tag dxs.med.11 cyss.high.1.ny glns.n.1tag secy.high.20 eno.high.20 cyss.low.1.ny prsa.n.1tag secy.low.1 eno.low.2.a cyss.med.1.ny nade.n.1tag secy.med.18 eno.low.2.b eno.high.1.ny pgsa.n.1tag trmd.low.2 fabg.high.20 eno.low.1.ny nrdb.n.1tag trmd.med.16 fabg.low.1 eno.med.1.ny gltx.n.1tag tyrs.med.18 fabg.med.18 era.high.1.ny lepb.n.1tag dxs.n.1tag isph.low.1 era.low.1.ny trmd.n.1tag dxs.n.2tag isph.med.10 era.med.1.ny grpe.n.1tag dxs.n.3tag isph.med.11 fabg.high.1.ny metk.n.1tag grpe.n.1tag lspa.high.20 fabg.low.1.ny infb.n.1tag grpe.n.2tag lspa.low.3 fabg.med.1.ny fmt.n.1tag grpe.n.3tag lspa.med.15 glns.high.1.ny secy.n.1tag rplo.n.1tag phet.high.20 glns.low.1.ny rplo.n.1tag rplo.n.2tag phet.low.2 glns.med.1.ny rpse.n.1tag rplo.n.3tag phet.med.14 gros.high.1.ny rpsn.n.1tag secy.n.1tag rplb.high.20 gros.low.1.ny rpln.n.1tag secy.n.2tag rplb.low.1.a gros.med.1.ny rpso.n.1tag secy.n.3tag rplb.low.1.b isph.high.1.ny rpld.n.1tag trmd.n.1tag rplj.low.4 isph.low.1.ny rplc.n.1tag trmd.n.2tag rplj.med.10 isph.med.1.ny fusa.n.1tag trmd.n.3tag rplj.med.13 lepb.high.1.ny rpoc.n.1tag rplm.low.2 lepb.low.1.ny ppa.n.1tag rplm.low.4 lepb.med.1.ny rplm.low.5 murd.high.1.ny rpls.low.1 murd.low.1.ny rpls.low.2 murd.med.1.ny rpmb.low.4 rplj.high.1.ny rpsd.low.1 rplj.high.2.ny rpsd.low.5 rplj.low.1.ny rpsk.low.8 rplj.low.2.ny rpsl.low.1 rplj.med.1.ny rpsl.low.2 rplj.med.2.ny rpsp.low.2 rplm.high.1.ny secy.high.20 rplm.low.1.ny secy.low.1 rplm.med.1.ny secy.med.18 rpls.high.1.ny topa.high.20 rpls.low.1.ny topa.low.5 rpls.med.1.ny topa.med.14 rpmd.high.1.ny trmd.low.2 rpmd.low.1.ny trmd.low.4 rpmd.med.1.ny trmd.med.16 rpoa.high.1.ny tyrs.low.1 rpoa.low.1.ny tyrs.med.18 rpoa.med.1.ny rpsb.high.1.ny sers.f213 was included in this pool 5
6 rpsb.low.1.ny rpsb.med.1.ny rpsd.high.1.ny rpsd.low.1.ny rpsd.med.1.ny thrs.high.1.ny thrs.low.1.ny thrs.med.1.ny trmd.high.1.ny trmd.high.2.ny trmd.low.1.ny trmd.low.2.ny trmd.med.1.ny trmd.med.2.ny Oligonucleotides within each pool were designed by a single or combination ("complex" pools) of strategies. Pool names are abbreviated as follows: Tolerant tyrosine (Y), tolerant non-tyrosine (NY), amino-terminal (N), complex 1 (C1), complex 2 (C2), and functional (F) pools. This file lists the oligonucleotides within six MAGE pools for the introduction of TAG codons within essential genes. Oligonucleotide names are defined in Supplementary Table 1. MAGE pool names: Tolerant tyrosine (Y), tolerant non-tyrosine (NY), aminoterminal (N), complex 1 (C1), complex 2 (C2), and functional (F). 6
7 Supplementary Table 3: Analysis of saa-dependence screens. MAGE Total Total Unique Unique Unique Unique Unique saa aars Pool a cycles screened dependent dependent 1TAG 2TAG 3TAG 4TAG pacf pacf-rs Y pacf pacf-rs NY n/a b n/a b n/a b n/a b pacf pacf-rs N pacf pacf-rs C pacf pacf-rs F n/a b n/a b n/a b n/a b pazf pazf-rs C n/a b n/a b n/a b n/a b pazf pcnf-rs Y pazf pcnf-rs N pazf pcnf-rs C pazf pcnf-rs F pif pif-rs C pif pif-rs F n/a b n/a b n/a b n/a b a Pool names are abbreviated as follows: Tolerant tyrosine (Y), tolerant non-tyrosine (NY), amino-terminal (N), complex 1 (C1), complex 2 (C2), and functional (F) pools. b We only quantitatively assess strains that were saa-dependent and cannot report all successful TAG incorporations that generated viable or nonviable noncontained clones. This file summarizes the results of screens for synthetic auxotrophs. Oligonucleotide pools are defined in Supplementary Table 2. Each aars is described in Supplementary Table
8 Supplementary Table 4: Conversion of aars specificity. Residue TyrRS (WT) pacf-rs pif-rs pazf-rs pcnf-rs 32 Y L L T L 107 E E S N D a 108 F F F F W 109 Q Q Q Q M 158 D G P P G 159 I C L L A 162 L R E Q L 286 D R R R R a The published pcnf-rs contains E107 but we pursued a D107 variant. >Methanocaldococcus jannaschii TyrRS MDEFEMIKRNTSEIISEEELREVLKKDEKSAYIGFEPSGKIHLGHYLQIKKMIDLQNA GFDIIILLADLHAYLNQKGELDEIRKIGDYNKKVFEAMGLKAKYVYGSEFQLDKDYTL NVYRLALKTTLKRARRSMELIAREDENPKVAEVIYPIMQVNDIHYLGVDVAVGGMEQR KIHMLARELLPKKVVCIHNPVLTGLDGEGKMSSSKGNFIAVDDSPEEIRAKIKKAYCP AGVVEGNPIMEIAKYFLEYPLTIKRPEKFGGDLTVNSYEELESLFKNKELHPMDLKNA VAEELIKILEPIRKRL This file contains the amino acid sequence of the M. jannaschii tyrosyl-trna synthetase used in this study. Residues changed relative to wild type (WT) TyrRS to convert the synthetic amino acid (saa) specificity, are highlighted. pcnf-rs is the relevant aars in all pazf auxotrophs listed in Supplementary Table
9 Supplementary Table 5: Summary of 22 essential genes containing TAG codons in saa-dependent strains. Essential gene(s) dxs ftsi metk murd pgsa can GO Annotation (Biological process) GO: (ubiquinone biosynthetic process) GO: (pyridoxine biosynthetic process) GO: (thiamine biosynthetic process) GO: (isoprenoid biosynthetic process) GO: ( terpenoid biosynthetic process) GO: (1-deoxy-D-xylulose 5-phosphate biosynthetic process) GO: (response to drug) GO: (cell division) GO: (cell cycle) GO: (regulation of cell shape) GO: (peptidoglycan biosynthetic process) GO: (S-adenosylmethionine biosynthetic process) GO: (one-carbon metabolic process) GO: (cell cycle) GO: (regulation of cell shape) GO: (biosynthetic process) GO: (peptidoglycan biosynthetic process) GO: (cell division) GO: (lipid metabolic process) GO: (phosphatidylglycerol biosynthetic process) GO: (phospholipid biosynthetic process) GO: (carbon utilization) GO Annotation (Molecular function) GO: (1-deoxy-D-xylulose-5-phosphate synthase activity) GO: (thiamine pyrophosphate binding)] GO: (magnesium ion binding) GO: (catalytic activity) GO: (transferase activity) GO: (metal ion binding) GO: (protein binding) GO: (penicillin binding) GO: (peptidoglycan glycosyltransferase activity) GO: (catalytic activity) GO: (transferase activity) GO: (transferase activity, transferring glycosyl groups) GO: (magnesium ion binding) GO: (methionine adenosyltransferase activity) GO: (protein binding) GO: (potassium ion binding) GO: (identical protein binding) GO: (nucleotide binding) GO: (ATP binding) GO: (transferase activity) GO: (metal ion binding) GO: (UDP-N-acetylmuramoylalanine-D-glutamate ligase activity) GO: (nucleotide binding) GO: (ATP binding) GO: (ligase activity) GO: (CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase activity) GO: (transferase activity) GO: (phosphotransferase activity, for other substituted phosphate groups) GO: (carbonate dehydratase activity) GO: (zinc ion binding) GO: (lyase activity) GO: (metal ion binding) dnax not found not found fabg GO: (fatty acid biosynthetic process) GO: (lipid biosynthetic process) GO: (biotin biosynthetic process) GO: (fatty acid elongation) GO: (lipid metabolic process) GO: (fatty acid metabolic process) GO: (oxidation-reduction process) isph lptd lspa phet rpoa secy sers topa tyrs GO: (ubiquinone biosynthetic process) GO: (isoprenoid biosynthetic process) GO: (terpenoid biosynthetic process) GO: (isopentenyl diphosphate biosynthetic process, methylerythritol 4-phosphate pathway) GO: (dimethylallyl diphosphate biosynthetic process) GO: (oxidation-reduction process) GO: (response to organic substance) GO: (lipopolysaccharide transport) GO: (Gram-negative-bacterium-type cell outer membrane assembly) GO: (membrane organization) GO: (signal peptide processing) GO: (proteolysis) GO: (translation) GO: (phenylalanyl-trna aminoacylation) GO: (trna processing) GO: (transcription, DNA-templated) GO: (intracellular protein transport) GO: (protein transport by the Sec complex) GO: (intracellular protein transmembrane transport) GO: (protein targeting) GO: (transpor) GO: (protein transport) GO: (seryl-trna aminoacylation) GO: (selenocysteine biosynthetic process) GO: (translation) GO: (trna aminoacylation for protein translation) GO: (selenocysteinyl-trna(sec) biosynthetic process) GO: (DNA topological change) GO: (DNA strand elongation) GO: (translation) GO: (trna aminoacylation for protein translation) GO: (tyrosyl-trna aminoacylation) GO: (3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity) GO: (identical protein binding) GO: (NADP binding) GO: (oxidoreductase activity) GO: (metal ion binding) GO: (NAD binding) GO: (hydroxymethylbutenyl pyrophosphate reductase activity) GO: (oxidoreductase activity) GO: (metal ion binding) GO: (iron-sulfur cluster binding) GO: (3 iron, 4 sulfur cluster binding) GO: (4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase activity) GO: (protein binding) GO: (peptidase activity) GO: (aspartic-type endopeptidase activity) GO: (hydrolase activity) GO: (protein binding) GO: (trna binding) GO: (nucleotide binding) GO: (magnesium ion binding) GO: (RNA binding) GO: (aminoacyl-trna ligase activity) GO: (phenylalanine-trna ligase activity) GO: (ATP binding) GO: (ligase activity) GO: (metal ion binding) GO: (protein binding) GO: (zinc ion binding) GO: (DNA binding) GO: (DNA-directed RNA polymerase activity) GO: (transferase activity) GO: (nucleotidyltransferase activity) GO: (protein dimerization activity) GO: (protein binding) GO: (magnesium ion binding) GO: (serine-trna ligase activity) GO: (identical protein binding) GO: (nucleotide binding) GO: (aminoacyl-trna ligase activity) GO: (ATP binding) GO: (ligase activity) GO: (DNA topoisomerase activity) GO: (DNA topoisomerase type I activity) GO: (nucleotide binding) GO: (magnesium ion binding) GO: (DNA binding) GO: (DNA topoisomerase type II (ATP-hydrolyzing) activity) GO: (ATP binding) GO: (isomerase activity) GO: (metal ion binding) GO: (nucleotide binding) GO: (RNA binding) GO: (aminoacyl-trna ligase activity) GO: (tyrosine-trna ligase activity) GO: (ATP binding) GO: (ligase activity) 9
10 dnaa fusa glyq lnt murg GO: (DNA replication) GO: (regulation of DNA replication) GO: (DNA replication initiation) GO: (transcription, DNA-templated) GO: (regulation of transcription, DNA-templated) GO: (negative regulation of DNA replication) GO: (GTP catabolic process) GO: (translation) GO: (translational elongation) GO: (translation) GO: (glycyl-trna aminoacylation) GO: (lipoprotein biosynthetic process) GO: (nitrogen compound metabolic process) GO: (peptidoglycan biosynthetic process) GO: (cell cycle) GO: (regulation of cell shape) GO: (UDP-N-acetylgalactosamine biosynthetic process) GO: (lipid glycosylation) GO: (cell division) GO: (DNA binding) GO: (DNA replication origin binding) GO: (protein binding) GO: (sequence-specific DNA binding) GO: (nucleotide binding) GO: (ATP binding) GO: (nucleoside-triphosphatase activity) GO: (nucleotide binding) GO: (translation elongation factor activity) GO: (GTPase activity) GO: (GTP binding) GO: (nucleotide binding) GO: (aminoacyl-trna ligase activity) GO: (glycine-trna ligase activity) GO: (ATP binding) GO: (ligase activity) GO: (N-acyltransferase activity) GO: (transferase activity) GO: (transferase activity, transferring acyl groups) GO: (hydrolase activity) GO: (hydrolase activity, acting on carbon-nitrogen [but not peptide] bonds) GO: (undecaprenyldiphospho-muramoylpentapeptide beta-n-acetylglucosaminyltransferase activity) GO: (transferase activity) GO: (transferase activity, transferring glycosyl groups) GO: (transferase activity, transferring hexosyl groups) GO: (carbohydrate binding) GO: (UDP-N-acetyl-D-glucosamine:N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6- diaminopimelyl-d-alanyl-d-alanine-diphosphoundecaprenol 4-beta-N-acetylglucosaminlytransferase activity) This file lists essential genes harboring TAG codons in synthetic auxotrophs from this study and associated GO annotations. 10
11 Supplementary Table 6: Summary of escape frequencies and doubling times for 60 saa-dependent strains. Nickname Synthetic amino acid Essential protein(s) TAG location(s) Escape frequency (EF) Doubling time (minutes) a (Day 1) 1mM saa 5mM saa rec.α pacf, α n/a n/a n/a ± 0.19 DnaX.Y113α pacf, α DnaX Y E-07 ± 1.89E ± 0.60 DnaX.Y113α' pacf, α DnaX Y E-07 ± 5.09E ± 0.73.Y54α pacf, α Y E-06 ± 1.16E ± 0.11 SecY.Y122α pacf, α SecY Y E-05 ± 8.38E ± 0.27 SecY.Y309α pacf, α SecY Y E-04 ± 3.15E ± 0.39 Dxs.αN pacf, α Dxs N-term 1.85E-06 ± 5.65E ± 0.15 FtsI.αN pacf, α FtsI N-term 1.07E-05 ± 6.27E ± 0.03 MurD.αN pacf, α MurD N-term 2.97E-05 ± 2.88E ± 0.41 rec.α.db.8 pacf, α TopA Y E-04 ± 4.45E ± 0.37 Y28 rec.β pif, β n/a n/a n/a ± 0.46 DnaX.Y113β pif, β DnaX Y E-07 ± 5.31E ± 0.68.Y54β pif, β Y E-05 ± 5.57E ± 1.52 SecY.Y122β pif, β SecY Y E-06 ± 6.47E ± 0.36 rec.β.db.9 pif, β DnaX Y E-08 ± 9.52E ± 0.29 Y54 rec.β.db.9' pif, β DnaX Y E-09 ± 1.62E ± 0.28 Y54 rec.β.dc.11 pif, β DnaX Y113 Y E-09 ± 3.61E ± 0.19 Dxs N-term rec.β.dc.11' rec.β.dc.12 rec.β.dc.12' rec.β.dc.12'.δty pif, β pif, β pif, β pif, β DnaX Dxs DnaX SecY DnaX SecY DnaX SecY Y113 Y54 N-term Y113 Y54 Y122 Y113 Y54 Y122 Y113 Y54 Y E-09 ± 4.90E ± E-09 ± 1.99E ± E-09 ± 3.69E ± 0.65 < 4.85E ± 0.70 rec.γ n/a n/a n/a ± ± 0.13 Can.Y83γ Can Y E-06 ± 7.90E ± 0.27 Can.Y99γ Can Y E-05 ± 3.84E ± 0.28 DnaX.Y113γ DnaX Y E-06 ± 2.08E ± 0.36 FabG.Y242γ FabG Y E-04 ± 6.96E ± 0.14 LptD.Y212γ LptD Y212 LptD.Y354γ LptD Y354 SecY.Y122γ SecY Y E-06 ± 5.28E ± 0.66 TyrS.Y127γ TyrS Y E-06 ± 1.75E ± 0.27 Dxs.γN Dxs N-term 3.41E-06 ± 2.10E ± 0.54 FtsI.γN FtsI N-term 1.53E-05 ± 1.15E ± 0.13 MurD.γN MurD N-term 4.24E-05 ± 2.57E ± 0.54 PgsA.γN PgsA N-term 1.36E-05 ± 3.22E ± 1.34 DnaA.W6γ DnaA W6 5.14E-07 ± 1.69E ± ± 0.28 FusA.F32γ FusA F E-03 ± 4.14E ± ±
12 GlyQ.Y226γ GlyQ Y E-05 ± 3.10E ± ± 1.19 Lnt.Y388γ Lnt Y E-06 ± 3.19E ± ± 0.06 MurG.F243γ MurG F E-07 ± 4.00E ± ± 0.52 SerS.F213γ SerS F E-06 ± 8.75E ± ± 0.36 rec.γ.db.16 PgsA N-term 9.87E-07 ± 4.24E ± 1.02 MetK rec.γ.db.17 SecY Y E-06 ± 1.14E ± 1.13 Y309 rec.γ.dc.18 rec.γ.dd.20 rec.γ.db.22 TopA SecY122 Dxs PheT IspH SecY PheT rec.γ.db.23 PheT rec.γ.dc.24 TopA Dxs rec.γ.db.26 DnaX rec.γ.db.28 LptD rec.γ.db.29 LptD rec.γ.db.31 LptD rec.γ.db.38 rec.γ.db.39 rec.γ.db.40 rec.γ.db.41 rec.γ.db.41' rec.γ.db.42 rec.γ.db.43 rec.γ.db.44 rec.γ.db.45 rec.γ.dc.46 rec.γ.dc.46' rec.γ.dc.46'.δty FusA RpoA FusA MurG DnaA GlyQ DnaA MurG DnaA MurG DnaA FusA DnaA SerS DnaA RpoA Lnt MurG DnaA MurG SerS DnaA MurG SerS DnaA MurG SerS a Average doubling times are shown for growth in permissive media; n=3 technical replicates; error is ±s.d. Y728 Y122 N-term Y28 Y50 Y615 Y142 Y309 Y615 Y558 Y601 Y728 Y28 N-term Y113 Y328 Y212 Y354 Y458 Y514 Y212 Y244 F32 Y152 F32 F243 W6 Y226 W6 F243 W6 F243 W6 F32 W6 F213 W6 Y152 Y388 F243 W6 F243 F213 W6 F243 F213 W6 F243 F E-05 ± 2.65E ± E-05 ± 5.70E ± E-05 ± 1.53E ± E-06 ± 3.73E ± E-06 ± 2.10E ± E-07 ± 8.69E ± E-09 ± 1.02E ± ± E ± ± E-09 ± 6.53E ± ± 0.10 < 7.88E ± ± 0.30 < 4.36E ± ± 0.23 < 6.3E ± ± 0.18 Summary of all synthetic auxotrophs. Average escape frequencies (EFs) refer to day 1, n=3 technical replicates, ±s.d. Representative EFs are shown in some cases (see methods). Average doubling times refer to growth in permissive media (0.2% L-arabinose & saa at 1 mm or 5 mm), n=3 technical replicates, ±s.d. 12
13 Supplementary Table 7: Escape mechanisms of MutS-deficient strains with one essential TAG codon. Escape mutant Essential gene Causative SNP a Causative SNP location b Relative fitness in media without saa c SecY.Y122α.E1 secy cag!cgg RpsD.Q54R 0.68 ± SecY.Y122α.E2 secy tag!cag SecY.α122Q 0.97 ± SecY.Y122α.E3 secy ctg!cta glnv!supe 1.00 ± DnaX.Y113α.E1 dnax tag!tgg DnaX.α113W 0.97 ± DnaX.Y113α.E2 dnax tag!tgg DnaX.α113W 0.95 ± DnaX.Y113α.E3 dnax tag!tgg DnaX.α113W 0.94 ± a Causative SNP is capitalized within the codon or anticodon sequence, from 5' to 3', for protein and trna-encoded genes, respectively. b Location of the amino acid change is shown for protein-encoded genes and names conferred by anticodon mutations for trna-encoded genes. c Average ratio of doubling times are shown for contained ancestor to escape mutant grown in permissive and nonpermissive media, respectively; n=3 technical replicates; error is ±s.d. This file summarizes observed escape mechanisms and fitness of escape mutants (EMs) of synthetic auxotrophs with one TAG codon and nonfunctional mismatch repair (ΔmutS). Average doubling time ratio for the contained ancestor to the EM grown in permissive and nonpermissive media, respectively are shown; n=3 technical replicates, ±s.d. 13
14 Supplementary Table 8: Whole genome sequencing of MutS-deficient strains with one essential TAG codon. Strain Feature Start End WT Mut Gene Consequence AA change Product GO Annotation (Biological process) DnaX.Y113α.E1 indel TCA thrl UPSTREAM (-188) - thr operon leader peptide GO: (cellular amino acid biosynthetic process) GO: (threonine biosynthetic process) GO: (transcriptional attenuation) GO: (transcriptional attenuation by ribosome) DnaX.Y113α.E1 snp A G dnax NON_SYNONYMOUS_CODING α/w DNA polymerase III/DNA elongation factor III, τ and γ subunits GO: (DNA replication) DnaX.Y113α.E1 snp T G mdfa START_REMOVED M/* multidrug efflux system protein DnaX.Y113α.E1 snp C T mdfa STOP_GAINED Q/* multidrug efflux system protein DnaX.Y113α.E1 snp C T ybji NON_SYNONYMOUS_CODING M/I FMN and erythrose-4-p phosphatase GO: (dephosphorylation) GO: (FMN catabolic process) lipoprotein required for formation of O-antigen capsule, predicted GO: (carbohydrate transport) GO: (ion transport) DnaX.Y113α.E1 snp C T gfce SYNONYMOUS_CODING A exopolysaccharide export protein GO: (polysaccharide transport) DnaX.Y113α.E1 snp C G stfp NON_SYNONYMOUS_CODING L/V uncharacterized protein from lambdoid prophage e14 region Not found SYNONYMOUS_CODING, Qin prophage; predicted tail fiber assembly protein (TfaQ), predicted tfaq: Not found ydfj: DnaX.Y113α.E1 snp C T tfaq, ydfj P, G/R NON_SYNONYMOUS_CODING transporter (YdfJ) GO: (potassium ion transport) DnaX.Y113α.E1 snp A G yedy NON_SYNONYMOUS_CODING D/G membrane-anchored, periplasmic TMAO, DMSO reductase GO: (oxidation reduction process) DnaX.Y113α.E1 indel G GC yfjm UPSTREAM (-11) - CP4-57 prophage, predicted protein Not found DnaX.Y113α.E1 snp C T yjjb SYNONYMOUS_CODING A conserved inner membrane protein Not found DnaX.Y113α.E1 snp TTT CAG thrl UPSTREAM (-194) - thr operon leader peptide GO: (cellular amino acid biosynthetic process) GO: (threonine biosynthetic process) GO: (transcriptional attenuation) GO: (transcriptional attenuation by ribosome) GO: (cellular amino acid biosynthetic process) GO: (threonine biosynthetic process) DnaX.Y113α.E2 indel 3 3 CT TATTTTTCAGCT thrl UPSTREAM (-187) - thr operon leader peptide GO: (transcriptional attenuation) GO: (transcriptional attenuation by ribosome) DnaX.Y113α.E2 snp A G dnax NON_SYNONYMOUS_CODING α/w DNA polymerase III/DNA elongation factor III, τ and γ subunits GO: (DNA replication) GO: (dephosphorylation) GO: (FMN catabolic process) DnaX.Y113α.E2 snp C T ybji NON_SYNONYMOUS_CODING M/I FMN and erythrose-4-p phosphatase DnaX.Y113α.E2 snp C G stfp NON_SYNONYMOUS_CODING L/V uncharacterized protein from lambdoid prophage e14 region Not found DnaX.Y113α.E2 snp C G stfe WITHIN_NON_CODING_GENE - pseudogene GO: (modulation by virus of host morphology or physiology) GO: (viral entry into host cell) GO: (virion attachment to host cell) DnaX.Y113α.E2 indel G GC yfjm UPSTREAM (-11) - CP4-57 prophage, predicted protein Not found DnaX.Y113α.E2 snp A G pgk SYNONYMOUS_CODING R phosphoglycerate kinase GO: (glycolysis) GO: (phosphorylation) DnaX.Y113α.E2 snp G A mrca SYNONYMOUS_CODING E fused penicillin-binding protein 1a: murein transglycosylase/murein GO: (negative regulation of cytolysis) GO: (peptidoglycan biosynthetic process) transpeptidase GO: (regulation of cell shape) DnaX.Y113α.E2 snp C T yjjb SYNONYMOUS_CODING A conserved inner membrane protein Not found GO: (cellular amino acid biosynthetic process) GO: (threonine biosynthetic process) DnaX.Y113α.E3 indel 2 2 GC TATTTTTCAGC thrl UPSTREAM (-188) - thr operon leader peptide GO: (transcriptional attenuation) GO: (transcriptional attenuation by ribosome) DnaX.Y113α.E3 snp A G dnax NON_SYNONYMOUS_CODING α/w DNA polymerase III/DNA elongation factor III, τ and γ subunits GO: (DNA replication) GO: (dephosphorylation) GO: (FMN catabolic process) DnaX.Y113α.E3 snp C T ybji NON_SYNONYMOUS_CODING M/I FMN and erythrose-4-p phosphatase DnaX.Y113α.E3 snp C G stfp NON_SYNONYMOUS_CODING L/V uncharacterized protein from lambdoid prophage e14 region Not found DnaX.Y113α.E3 snp G A torz SYNONYMOUS_CODING G trimethylamine N-oxide reductase system III, catalytic subunit GO: (cellular respiration) GO: (oxidation-reduction process) fused enoyl-coa hydratase and epimerase and isomerase/3- GO: (fatty acid beta-oxidation) GO: (fatty acid metabolic process) GO: (lipid catabolic process) DnaX.Y113α.E3 snp G A fadj NON_SYNONYMOUS_CODING T/I hydroxyacyl-coa dehydrogenase GO: (lipid metabolic process) GO: (oxidation-reduction process) DnaX.Y113α.E3 snp T C hyfd NON_SYNONYMOUS_CODING L/P hydrogenase 4, membrane subunit GO: (ATP synthesis coupled electron transport) GO: (oxidation-reduction process) DnaX.Y113α.E3 indel G GC yfjm UPSTREAM (-11) - CP4-57 prophage, predicted protein Not found DnaX.Y113α.E3 snp C T nrdd NON_SYNONYMOUS_CODING A/T anaerobic ribonucleoside-triphosphate reductase GO: (DNA replication) GO: (nucleobase-containing small molecule interconversion) GO: (oxidation-reduction process) DnaX.Y113α.E3 snp C T yjjb SYNONYMOUS_CODING A conserved inner membrane protein Not found insn: GO: (transposition, DNA-mediated) GO: (transposition) insi1: SecY.Y122α.E1 snp T C insn, insi1 NON_SYNONYMOUS_CODING, Y/H, N pseudogene (IsnN), IS30 transposase (InsI1) SYNONYMOUS_CODING GO: (transposition, DNA-mediated) GO: (DNA integration) GO: (transposition) GO: (copper ion homeostasis) GO: (detoxification of copper ion) GO: (response to copper ion) SecY.Y122α.E1 snp C T cusf NON_SYNONYMOUS_CODING P/L periplasmic copper- and silver-binding protein GO: (response to silver ion) GO: (response to zinc ion) SecY.Y122α.E1 indel G rhsc FRAMESHIFT_CODING - rhsc element core protein RshC Not found DNA-dependent ATPase involved in processing recombination SecY.Y122α.E1 snp G A rara NON_SYNONYMOUS_CODING V/I intermediates at replication forks GO: (DNA-dependent DNA replication) SecY.Y122α.E1 snp G A foca SYNONYMOUS_CODING L formate channel GO: (formate transport) SecY.Y122α.E1 snp G A wzc NON_SYNONYMOUS_CODING P/S protein-tyrosine kinase GO: (capsule polysaccharide biosynthetic process) GO: (colanic acid biosynthetic process) GO: (lipopolysaccharide biosynthetic process) GO: (peptidyl-tyrosine autophosphorylation) GO: (peptidyl-tyrosine phosphorylation) GO: (phosphorylation) GO: (polysaccharide biosynthetic process) GO: (regulation of catalytic activity) SecY.Y122α.E1 indel G - sstt FRAMESHIFT_CODING - pseudogene GO: (amino acid transmembrane transport) GO: (amino acid transport) GO: (dicarboxylic acid transport) GO: (serine transport) GO: (threonine transport) SecY.Y122α.E1 snp C T elbb NON_SYNONYMOUS_CODING R/H isoprenoid biosynthesis protein with amidotransferase-like domain GO: (isoprenoid biosynthetic process) GO: (positive regulation of isoprenoid metabolic process) 14
Supporting Information
 Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Glyma10g15850 aagggatccattctggaaccatatcttgctgtg ttgggtacccttggatgcaggatgacacg AtMKK6, AT5g56580
 Supplemental table 1. Soybean MAPK, MAPKK, and MAPKKK genes and primers for VIGS cloning Gene_ID Forward Primer Reverse Primer Arabidopsis Ortholog Glyma04g03210 aagggatcctgcagcaagaaccagttcat ttgggtaccctaatggatgcgcattaggataaag
Supplemental table 1. Soybean MAPK, MAPKK, and MAPKKK genes and primers for VIGS cloning Gene_ID Forward Primer Reverse Primer Arabidopsis Ortholog Glyma04g03210 aagggatcctgcagcaagaaccagttcat ttgggtaccctaatggatgcgcattaggataaag
Supplementary Table 2. Gossypol-induced Decrease of Levels of Reduced Cysteine-containing Peptides
 Supplementary Table 2. Gossypol-induced Decrease of Levels of Reduced Cysteine-containing Peptides Accession Description Sequences of Cys-containing peptides IPI00021263 14-3-3 protein zeta/delta DICNDVLSLLEK
Supplementary Table 2. Gossypol-induced Decrease of Levels of Reduced Cysteine-containing Peptides Accession Description Sequences of Cys-containing peptides IPI00021263 14-3-3 protein zeta/delta DICNDVLSLLEK
Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley M. Lemon, Lawrence M. Pfeffer, Kui Li
 Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Architecture of the Trypanosome RNA Editing Accessory Complex, MRB1
 Architecture of the Trypanosome RNA Editing Accessory Complex, MRB1 Michelle L. Ammerman, Kurtis M. Downey, Hassan Hashimi, John C. Fisk, Danielle L. Tomasello, Drahomíra Faktorová, Lucie Hanzálková, Tony
Architecture of the Trypanosome RNA Editing Accessory Complex, MRB1 Michelle L. Ammerman, Kurtis M. Downey, Hassan Hashimi, John C. Fisk, Danielle L. Tomasello, Drahomíra Faktorová, Lucie Hanzálková, Tony
Modular Optimization of Hemicellulose-utilizing Pathway in. Corynebacterium glutamicum for Consolidated Bioprocessing of
 [Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
[Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
Supplementary Table 1. Cystometric parameters in sham-operated wild type and Trpv4 -/- rats during saline infusion and
 WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
kpis(ppk20) kpis(ppk46)
 Table S1. C. elegans strains used in this study Strains carrying transgenes Strain Transgene Genotype Reference IT213 tcer-1 prom:tcer-1orf:gfp:tcer-1 3'utr unc-119(ed3) III; kpis(ppk6) This study IT283
Table S1. C. elegans strains used in this study Strains carrying transgenes Strain Transgene Genotype Reference IT213 tcer-1 prom:tcer-1orf:gfp:tcer-1 3'utr unc-119(ed3) III; kpis(ppk6) This study IT283
Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali. Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang*
 Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Flowering time. Col C24 Cvi C24xCol C24xCvi ColxCvi
 Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Tutorial 1 The Central Dogma of molecular biology
 oday DN RN rotein utorial 1 he entral Dogma of molecular biology Information flow in genetics:» ranscription» ranslation» Making sense of genomic information Information content in DN - Information content
oday DN RN rotein utorial 1 he entral Dogma of molecular biology Information flow in genetics:» ranscription» ranslation» Making sense of genomic information Information content in DN - Information content
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing MFMER slide-1
 Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Suppl. Materials. Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids. Germany
 Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Mapping Sequencing Reads to a Reference Genome
 Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo
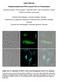
 Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
SOLiD Technology. library preparation & Sequencing Chemistry (sequencing by ligation!) Imaging and analysis. Application specific sample preparation
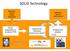 SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
tccattaattcgacagaccagagttaaataatccttgtatgccattgtgatcacatctacagttcagattttgtatttca
 Figure Legends for Supplementary Materials: Fig. 1. Nucleotide sequence of the zebrafish lumican (zlum) gene. Exons are indicated by uppercase letters, and introns are indicated by lowercase letters. The
Figure Legends for Supplementary Materials: Fig. 1. Nucleotide sequence of the zebrafish lumican (zlum) gene. Exons are indicated by uppercase letters, and introns are indicated by lowercase letters. The
Phenotype. Genotype. It is like any other experiment! What is a bioinformatics experiment? Remember the Goal. Infectious Disease Paradigm
 It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
Akt1 Akt kinase activity Creb signaling CCTTACAGCCCTCAAGTACTCATTC GGCGTACTCCATGACAAAGCA Arc Actin binding
 Additional File 1 (Table S1): Description and primer sequences of 80 candidate genes tested as potential synaptic plasticity formation genes in qrt-pcr array. Abbreviations: LTP: long-term potentiation,
Additional File 1 (Table S1): Description and primer sequences of 80 candidate genes tested as potential synaptic plasticity formation genes in qrt-pcr array. Abbreviations: LTP: long-term potentiation,
Supplementary Figure 1
 Supplementary Figure 1 Plot of delta-afe of sequence variants detected between resistant and susceptible pool over the genome sequence of the WB42 line of B. vulgaris ssp. maritima. The delta-afe values
Supplementary Figure 1 Plot of delta-afe of sequence variants detected between resistant and susceptible pool over the genome sequence of the WB42 line of B. vulgaris ssp. maritima. The delta-afe values
Expression analysis of PIN genes in root tips and nodules of Lotus japonicus
 Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and
 1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
A cell-based screening system for RNA Polymerase I inhibitors
 Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
University of Bristol - Explore Bristol Research
 Tan, B. G., Wellesley, F. C., Savery, N. J., & Szczelkun, M. D. (2016). Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity.
Tan, B. G., Wellesley, F. C., Savery, N. J., & Szczelkun, M. D. (2016). Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity.
Supporting Information
 Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
The beet R locus encodes a new cytochrome P450 required for red. betalain production.
 The beet R locus encodes a new cytochrome P450 required for red betalain production. Gregory J. Hatlestad, Rasika M. Sunnadeniya, Neda A. Akhavan, Antonio Gonzalez, Irwin L. Goldman, J. Mitchell McGrath,
The beet R locus encodes a new cytochrome P450 required for red betalain production. Gregory J. Hatlestad, Rasika M. Sunnadeniya, Neda A. Akhavan, Antonio Gonzalez, Irwin L. Goldman, J. Mitchell McGrath,
Bioinformatics: Blending. Biology and Computer Science
 Bioinformatics: Blending Biology and Computer Science MDNMSITNTPTSNDACLSIVHSLMCHRQ GGESETFAKRAIESLVKKLKEKKDELDSL ITAITTNGAHPSKCVTIQRTLDGRLQVAG RKGFPHVIYARLWRWPDLHKNELKHVK YCQYAFDLKCDSVCVNPYHYERVVSPGI DLSGLTLQSNAPSSMMVKDEYVHDFEG
Bioinformatics: Blending Biology and Computer Science MDNMSITNTPTSNDACLSIVHSLMCHRQ GGESETFAKRAIESLVKKLKEKKDELDSL ITAITTNGAHPSKCVTIQRTLDGRLQVAG RKGFPHVIYARLWRWPDLHKNELKHVK YCQYAFDLKCDSVCVNPYHYERVVSPGI DLSGLTLQSNAPSSMMVKDEYVHDFEG
T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma
 T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma Prescott G. Woodruff, M.D., M.P.H., Barmak Modrek, Ph.D., David F. Choy, B.S., Guiquan Jia, M.D., Alexander R. Abbas, Ph.D., Almut
T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma Prescott G. Woodruff, M.D., M.P.H., Barmak Modrek, Ph.D., David F. Choy, B.S., Guiquan Jia, M.D., Alexander R. Abbas, Ph.D., Almut
Forensic SNP Genotyping using Nanopore MinION Sequencing
 Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
Supplemental Information. Investigation of Penicillin Binding Protein. (PBP)-like Peptide Cyclase and Hydrolase
 Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
SUPPLEMENTAL DATA. Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli.
 SUPPLEMENTAL DATA Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli. B. Douzi, Y.R. Brunet, S. Spinelli, V. Lensi, P. Legrand, S. Blangy,
SUPPLEMENTAL DATA Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli. B. Douzi, Y.R. Brunet, S. Spinelli, V. Lensi, P. Legrand, S. Blangy,
Supporting Information
 Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Correlation & Linear Regression in SPSS
 Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
The role of aristolochene synthase in diphosphate activation
 Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Nevezze meg a jelölt csontot latinul! Name the bone marked! Nevezze meg a jelölt csont típusát! What is the type of the bone marked?
 1 Nevezze meg a jelölt csontot latinul! Name the bone marked! 2 Nevezze meg a jelölt csont típusát! What is the type of the bone marked? 3 Milyen csontállomány található a jelölt csont belsejében? What
1 Nevezze meg a jelölt csontot latinul! Name the bone marked! 2 Nevezze meg a jelölt csont típusát! What is the type of the bone marked? 3 Milyen csontállomány található a jelölt csont belsejében? What
PETER PAZMANY CATHOLIC UNIVERSITY Consortium members SEMMELWEIS UNIVERSITY, DIALOG CAMPUS PUBLISHER
 SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
Sebastián Sáez Senior Trade Economist INTERNATIONAL TRADE DEPARTMENT WORLD BANK
 Sebastián Sáez Senior Trade Economist INTERNATIONAL TRADE DEPARTMENT WORLD BANK Despite enormous challenges many developing countries are service exporters Besides traditional activities such as tourism;
Sebastián Sáez Senior Trade Economist INTERNATIONAL TRADE DEPARTMENT WORLD BANK Despite enormous challenges many developing countries are service exporters Besides traditional activities such as tourism;
First experiences with Gd fuel assemblies in. Tamás Parkó, Botond Beliczai AER Symposium 2009.09.21 25.
 First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
Fruits Amount Minerals Contained Vitamins Contained. Calcium -9.5 mg Phosphorus - 95mg 9.5 Magnesium -7 mg
 TERMÉSEK NÉHÁNY FONTOSABB GYÜMÖLCS ELEMTARTALMA ÉS? VITAMINTARTALMA Note that only those nutrients which appear in significant quantities are listed. For more detailed information, please visit the United
TERMÉSEK NÉHÁNY FONTOSABB GYÜMÖLCS ELEMTARTALMA ÉS? VITAMINTARTALMA Note that only those nutrients which appear in significant quantities are listed. For more detailed information, please visit the United
Statistical Dependence
 Statistical Dependence Petra Petrovics Statistical Dependence Deinition: Statistical dependence exists when the value o some variable is dependent upon or aected by the value o some other variable. Independent
Statistical Dependence Petra Petrovics Statistical Dependence Deinition: Statistical dependence exists when the value o some variable is dependent upon or aected by the value o some other variable. Independent
Genetikai panel kialakítása a hazai tejhasznú szarvasmarha állományok hasznos élettartamának növelésére
 Genetikai panel kialakítása a hazai tejhasznú szarvasmarha állományok hasznos élettartamának növelésére Dr. Czeglédi Levente Dr. Béri Béla Kutatás-fejlesztés támogatása a megújuló energiaforrások és agrár
Genetikai panel kialakítása a hazai tejhasznú szarvasmarha állományok hasznos élettartamának növelésére Dr. Czeglédi Levente Dr. Béri Béla Kutatás-fejlesztés támogatása a megújuló energiaforrások és agrár
Crash Course in Omics Terminology, Concepts & Data Types
 Crash Course in Omics Terminology, Concepts & Data Types 12 th Annual EuPathDB Workshop: 2017 Jessie Kissinger June 18, 2017 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB
Crash Course in Omics Terminology, Concepts & Data Types 12 th Annual EuPathDB Workshop: 2017 Jessie Kissinger June 18, 2017 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB
CLUSTALW Multiple Sequence Alignment
 Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
STUDENT LOGBOOK. 1 week general practice course for the 6 th year medical students SEMMELWEIS EGYETEM. Name of the student:
 STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
Membrántranszport. Gyógyszerész előadás Dr. Barkó Szilvia
 Membrántranszport Gyógyszerész előadás 2017.04.10 Dr. Barkó Szilvia Sejt membránok A sejtmembrán funkciói Védelem Kommunikáció Molekulák importja és exportja Sejtmozgás Általános szerkezet Lipid kettősréteg
Membrántranszport Gyógyszerész előadás 2017.04.10 Dr. Barkó Szilvia Sejt membránok A sejtmembrán funkciói Védelem Kommunikáció Molekulák importja és exportja Sejtmozgás Általános szerkezet Lipid kettősréteg
Átlagtermés és rekordtermés 8 növénykultúrában
 Átlagtermés és rekordtermés 8 növénykultúrában Növény Rekord termés * Átlag termés * Veszteség típusa Biotikus Abiotikus Abiotikus stressz okozta kiesés (% a rekord terméshez képest) Kukorica 19300 4600
Átlagtermés és rekordtermés 8 növénykultúrában Növény Rekord termés * Átlag termés * Veszteség típusa Biotikus Abiotikus Abiotikus stressz okozta kiesés (% a rekord terméshez képest) Kukorica 19300 4600
discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo
 discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo Download and install discosnp demo - Peterlongo Pierre 3 Download web page: github.com/gatb/discosnp Chose latest release (2.2.10 today) discosnp
discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo Download and install discosnp demo - Peterlongo Pierre 3 Download web page: github.com/gatb/discosnp Chose latest release (2.2.10 today) discosnp
On The Number Of Slim Semimodular Lattices
 On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
A rosszindulatú daganatos halálozás változása 1975 és 2001 között Magyarországon
 A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
Characteristics and categorization of transportation organizations
 Characteristics and categorization of transportation organizations Organisational structure Activity (function) structure functional unit organisational unit sub-system input, stored, output information
Characteristics and categorization of transportation organizations Organisational structure Activity (function) structure functional unit organisational unit sub-system input, stored, output information
Összefoglalás. Summary. Bevezetés
 A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
HALLGATÓI KÉRDŐÍV ÉS TESZT ÉRTÉKELÉSE
 HALLGATÓI KÉRDŐÍV ÉS TESZT ÉRTÉKELÉSE EVALUATION OF STUDENT QUESTIONNAIRE AND TEST Daragó László, Dinyáné Szabó Marianna, Sára Zoltán, Jávor András Semmelweis Egyetem, Egészségügyi Informatikai Fejlesztő
HALLGATÓI KÉRDŐÍV ÉS TESZT ÉRTÉKELÉSE EVALUATION OF STUDENT QUESTIONNAIRE AND TEST Daragó László, Dinyáné Szabó Marianna, Sára Zoltán, Jávor András Semmelweis Egyetem, Egészségügyi Informatikai Fejlesztő
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites
 Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
Számlakezelés az ELO DocXtraktor modullal
 ELOECMSzakmai Kongresszus2013 Számlakezelés az ELO DocXtraktor modullal Kovács Eszter Kovacs.eszter@pentatrade.hu Projekt bemutatása A Cég Cégcsoport Éves árbevétel 140 mrd FT > 5 500 dolgozó ( 1 000 fı
ELOECMSzakmai Kongresszus2013 Számlakezelés az ELO DocXtraktor modullal Kovács Eszter Kovacs.eszter@pentatrade.hu Projekt bemutatása A Cég Cégcsoport Éves árbevétel 140 mrd FT > 5 500 dolgozó ( 1 000 fı
White Paper. Grounding Patch Panels
 White Paper Grounding Patch Panels Tartalom 1. Bevezető... 1 2. A földelés jelentőssége... 3 3. AC elosztó rendszer... 3 4. Földelési rendszerek... 3 4.1. Fa... 3 4.2. Háló... 5 5. Patch panel földelési
White Paper Grounding Patch Panels Tartalom 1. Bevezető... 1 2. A földelés jelentőssége... 3 3. AC elosztó rendszer... 3 4. Földelési rendszerek... 3 4.1. Fa... 3 4.2. Háló... 5 5. Patch panel földelési
USER MANUAL Guest user
 USER MANUAL Guest user 1 Welcome in Kutatótér (Researchroom) Top menu 1. Click on it and the left side menu will pop up 2. With the slider you can make left side menu visible 3. Font side: enlarging font
USER MANUAL Guest user 1 Welcome in Kutatótér (Researchroom) Top menu 1. Click on it and the left side menu will pop up 2. With the slider you can make left side menu visible 3. Font side: enlarging font
Biológus MSc. Molekuláris biológiai alapismeretek
 Biológus MSc Molekuláris biológiai alapismeretek A nukleotidok építőkövei A nukleotidok szerkezete Nukleotid = N-tartalmú szerves bázis + pentóz + foszfát N-glikozidos kötés 5 1 4 2 3 (Foszfát)észter-kötés
Biológus MSc Molekuláris biológiai alapismeretek A nukleotidok építőkövei A nukleotidok szerkezete Nukleotid = N-tartalmú szerves bázis + pentóz + foszfát N-glikozidos kötés 5 1 4 2 3 (Foszfát)észter-kötés
Construction of a cube given with its centre and a sideline
 Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
University of Bristol - Explore Bristol Research
 Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress Supplementary Figure S1.
 Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress Sheila I. Jensen 1*, Rebecca M. Lennen 1, Markus J. Herrgård 1, Alex T. Nielsen 1*
Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress Sheila I. Jensen 1*, Rebecca M. Lennen 1, Markus J. Herrgård 1, Alex T. Nielsen 1*
PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING PROPERTIES
 Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 371 379. PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING
Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 371 379. PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING
Étrend-kiegészítık és evidenciák mégis mit szedjek?
 Étrend-kiegészítık és evidenciák mégis mit szedjek? Dr. Lugasi Andrea Országos Élelmezés- és Táplálkozástudományi Intézet, Budapest Sport és Táplálkozás, 2009. november 11. Étrend-kiegészítı Mi az? Élelmiszer
Étrend-kiegészítık és evidenciák mégis mit szedjek? Dr. Lugasi Andrea Országos Élelmezés- és Táplálkozástudományi Intézet, Budapest Sport és Táplálkozás, 2009. november 11. Étrend-kiegészítı Mi az? Élelmiszer
Bérczi László tű. dandártábornok Országos Tűzoltósági Főfelügyelő
 The role of volunteer firefighter organizations, municipality and facility firefighter departments in the unified disaster management system of Hungary Bérczi László tű. dandártábornok Országos Tűzoltósági
The role of volunteer firefighter organizations, municipality and facility firefighter departments in the unified disaster management system of Hungary Bérczi László tű. dandártábornok Országos Tűzoltósági
A minimális sejt. Avagy hogyan alkalmazzuk a biológia több területét egy kérdés megválaszolására
 A minimális sejt Avagy hogyan alkalmazzuk a biológia több területét egy kérdés megválaszolására Anyagcsere Gánti kemoton elmélete Minimum sejt Top down: Meglevő szervezetek genomjából indulunk ki Bottom
A minimális sejt Avagy hogyan alkalmazzuk a biológia több területét egy kérdés megválaszolására Anyagcsere Gánti kemoton elmélete Minimum sejt Top down: Meglevő szervezetek genomjából indulunk ki Bottom
pjnc-pgluc Codon-optimized Gaussia luciferase (pgluc) Vector backbone: JM83 pjnc-pgluc is resistant to ampicillin and neomycin High or low copy:
 pjnc-pgluc Gene/Insert name: Codon-optimized Gaussia luciferase (pgluc) Vector backbone: pcmv-jnc Vector type: Mammalian cells Backbone size w/o insert (bp): 5375 Bacterial resistance: Ampicillin and neomycin
pjnc-pgluc Gene/Insert name: Codon-optimized Gaussia luciferase (pgluc) Vector backbone: pcmv-jnc Vector type: Mammalian cells Backbone size w/o insert (bp): 5375 Bacterial resistance: Ampicillin and neomycin
Oxacillin MIC µ g ml borderline oxacillin-resistant Staphylococcus aureus. oxacillin Staphylococcus aureus MRSA
 oxacillin Staphylococcus aureus MRSA 8 17 11 1 Oxacillin MIC 1248 µ gml borderline oxacillin-resistant Staphylococcus aureus BORSA79 NCCLS CLSIM100-S142004 cefoxitin disk oxacillin cefoxitin methicillin-resistant
oxacillin Staphylococcus aureus MRSA 8 17 11 1 Oxacillin MIC 1248 µ gml borderline oxacillin-resistant Staphylococcus aureus BORSA79 NCCLS CLSIM100-S142004 cefoxitin disk oxacillin cefoxitin methicillin-resistant
CISZTÁS FIBRÓZIS GYÓGYÍTHATÓ?!
 CISZTÁS FIBRÓZIS GYÓGYÍTHATÓ?! Ujhelyi Rita 2012. November Gyermekpulmológiai Konferencia Cisztás fibrózis Öröklődő betegség Nem gyógyítható Klorid ion transzport zavar Légutak és emésztőrendszert érinti
CISZTÁS FIBRÓZIS GYÓGYÍTHATÓ?! Ujhelyi Rita 2012. November Gyermekpulmológiai Konferencia Cisztás fibrózis Öröklődő betegség Nem gyógyítható Klorid ion transzport zavar Légutak és emésztőrendszert érinti
BKI13ATEX0030/1 EK-Típus Vizsgálati Tanúsítvány/ EC-Type Examination Certificate 1. kiegészítés / Amendment 1 MSZ EN 60079-31:2014
 (1) EK-TípusVizsgálati Tanúsítvány (2) A potenciálisan robbanásveszélyes környezetben történő alkalmazásra szánt berendezések, védelmi rendszerek 94/9/EK Direktíva / Equipment or Protective Systems Intended
(1) EK-TípusVizsgálati Tanúsítvány (2) A potenciálisan robbanásveszélyes környezetben történő alkalmazásra szánt berendezések, védelmi rendszerek 94/9/EK Direktíva / Equipment or Protective Systems Intended
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet Nonparametric Tests
 Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
AZ ÁRPA SZÁRAZSÁGTŰRÉSÉNEK VIZSGÁLATA: QTL- ÉS ASSZOCIÁCIÓS ANALÍZIS, MARKER ALAPÚ SZELEKCIÓ, TILLING
 Hagyomány és haladás a növénynemesítésben AZ ÁRPA SZÁRAZSÁGTŰRÉSÉNEK VIZSGÁLATA: QTL- ÉS ASSZOCIÁCIÓS ANALÍZIS, MARKER ALAPÚ SZELEKCIÓ, BÁLINT ANDRÁS FERENC 1, SZIRA FRUZSINA 1, ANDREAS BÖRNER 2, KERSTIN
Hagyomány és haladás a növénynemesítésben AZ ÁRPA SZÁRAZSÁGTŰRÉSÉNEK VIZSGÁLATA: QTL- ÉS ASSZOCIÁCIÓS ANALÍZIS, MARKER ALAPÚ SZELEKCIÓ, BÁLINT ANDRÁS FERENC 1, SZIRA FRUZSINA 1, ANDREAS BÖRNER 2, KERSTIN
Computational Neuroscience
 Computational Neuroscience Zoltán Somogyvári senior research fellow KFKI Research Institute for Particle and Nuclear Physics Supporting materials: http://www.kfki.hu/~soma/bscs/ BSCS 2010 Lengyel Máté:
Computational Neuroscience Zoltán Somogyvári senior research fellow KFKI Research Institute for Particle and Nuclear Physics Supporting materials: http://www.kfki.hu/~soma/bscs/ BSCS 2010 Lengyel Máté:
Crash Course in Omics Terminology, Concepts & Data Types
 Crash Course in Omics Terminology, Concepts & Data Types 13 th Annual EuPathDB Workshop Jessie Kissinger June 17th, 2018 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB GiardiaDB
Crash Course in Omics Terminology, Concepts & Data Types 13 th Annual EuPathDB Workshop Jessie Kissinger June 17th, 2018 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB GiardiaDB
Excel vagy Given-When-Then? Vagy mindkettő?
 TESZT & TEA BUDAPEST AGILE MEETUP Pénzügyi számítások automatizált agilis tesztelése: Excel vagy Given-When-Then? Vagy mindkettő? NAGY GÁSPÁR TechTalk developer coach Budapest, 2014 február 6. SpecFlow
TESZT & TEA BUDAPEST AGILE MEETUP Pénzügyi számítások automatizált agilis tesztelése: Excel vagy Given-When-Then? Vagy mindkettő? NAGY GÁSPÁR TechTalk developer coach Budapest, 2014 február 6. SpecFlow
Limitations and challenges of genetic barcode quantification
 Limitations and challenges of genetic barcode quantification Lars hielecke, im ranyossy, ndreas Dahl, Rajiv iwari, Ingo Roeder, Hartmut eiger, Boris Fehse, Ingmar lauche and Kerstin ornils SUPPLEMENRY
Limitations and challenges of genetic barcode quantification Lars hielecke, im ranyossy, ndreas Dahl, Rajiv iwari, Ingo Roeder, Hartmut eiger, Boris Fehse, Ingmar lauche and Kerstin ornils SUPPLEMENRY
Supplemental Information. RNase H1-Dependent Antisense Oligonucleotides. Are Robustly Active in Directing RNA Cleavage
 YMTHE, Volume 25 Supplemental Information RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus Xue-Hai Liang, Hong Sun, Joshua
YMTHE, Volume 25 Supplemental Information RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus Xue-Hai Liang, Hong Sun, Joshua
A magkémia alapjai. Kinetika. Nagy Sándor ELTE, Kémiai Intézet
 A magkémia alapjai Kinetika Nagy Sándor ELTE, Kémiai Intézet 09 The Radium Girls Festék világít Néhány egyszerű empirikus fogalomra teszünk egy pár triviális észrevételt. Egyetlen iterációban finomítjuk
A magkémia alapjai Kinetika Nagy Sándor ELTE, Kémiai Intézet 09 The Radium Girls Festék világít Néhány egyszerű empirikus fogalomra teszünk egy pár triviális észrevételt. Egyetlen iterációban finomítjuk
Analitikai megoldások IBM Power és FlashSystem alapokon. Mosolygó Ferenc - Avnet
 Analitikai megoldások IBM Power és FlashSystem alapokon Mosolygó Ferenc - Avnet Bevezető Legfontosabb elvárásaink az adatbázisokkal szemben Teljesítmény Lekérdezések, riportok és válaszok gyors megjelenítése
Analitikai megoldások IBM Power és FlashSystem alapokon Mosolygó Ferenc - Avnet Bevezető Legfontosabb elvárásaink az adatbázisokkal szemben Teljesítmény Lekérdezések, riportok és válaszok gyors megjelenítése
Cashback 2015 Deposit Promotion teljes szabályzat
 Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
ELOECMSzakmai Kongresszus2013
 ELOECMSzakmai Kongresszus2013 Keynote Horváth Szilvia Ügyvezető s.horvath@elo.com Cégünk rövid bemutatása 1871 Louis Leitz megalapítja első vállalatát 1995 Az első elektronikus Leitz dokumentumkezelő (ELOoffice)
ELOECMSzakmai Kongresszus2013 Keynote Horváth Szilvia Ügyvezető s.horvath@elo.com Cégünk rövid bemutatása 1871 Louis Leitz megalapítja első vállalatát 1995 Az első elektronikus Leitz dokumentumkezelő (ELOoffice)
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Correlation & Linear. Petra Petrovics.
 Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
DECLARATION OF PERFORMANCE No. GST REV 1.03 According to Construction Products Regulation EU No. 305/2011
 DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
FÖLDRAJZ ANGOL NYELVEN
 Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke
 A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke A dokumentum A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásában
A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke A dokumentum A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásában
KN-CP50. MANUAL (p. 2) Digital compass. ANLEITUNG (s. 4) Digitaler Kompass. GEBRUIKSAANWIJZING (p. 10) Digitaal kompas
 KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
klorid ioncsatorna az ABC (ATP Binding Casette) fehérjecsaládba tartozik, amelyek általánosságban részt vesznek a gyógyszerek olyan alapvetı
 Szegedi Tudományegyetem Farmakológiai és Farmakoterápiai Intézet Igazgató: Prof. Dr. Varró András 6720 Szeged, Dóm tér 12., Tel.: (62) 545-682 Fax: (62) 545-680 e-mail: varro.andras@med.u-szeged.hu Opponensi
Szegedi Tudományegyetem Farmakológiai és Farmakoterápiai Intézet Igazgató: Prof. Dr. Varró András 6720 Szeged, Dóm tér 12., Tel.: (62) 545-682 Fax: (62) 545-680 e-mail: varro.andras@med.u-szeged.hu Opponensi
Supplemental Table S1. Overview of MYB transcription factor genes analyzed for expression in red and pink tomato fruit.
 Supplemental Table S1. Overview of MYB transcription factor genes analyzed for expression in red and pink tomato fruit. MYB Primer pairs TC AtMYB Forward Reverse TC199266 MYB12 AGGCTCTTGGAGGTCGTTACC CAACTCTTTCCGCATCTCAATAATC
Supplemental Table S1. Overview of MYB transcription factor genes analyzed for expression in red and pink tomato fruit. MYB Primer pairs TC AtMYB Forward Reverse TC199266 MYB12 AGGCTCTTGGAGGTCGTTACC CAACTCTTTCCGCATCTCAATAATC
(11) Lajstromszám: E 005 557 (13) T2 EURÓPAI SZABADALOM SZÖVEGÉNEK FORDÍTÁSA
 !HU000007T2! (19) HU (11) Lajstromszám: E 00 7 (13) T2 MAGYAR KÖZTÁRSASÁG Magyar Szabadalmi Hivatal EURÓPAI SZABADALOM SZÖVEGÉNEK FORDÍTÁSA (21) Magyar ügyszám: E 03 026690 (22) A bejelentés napja: 03.
!HU000007T2! (19) HU (11) Lajstromszám: E 00 7 (13) T2 MAGYAR KÖZTÁRSASÁG Magyar Szabadalmi Hivatal EURÓPAI SZABADALOM SZÖVEGÉNEK FORDÍTÁSA (21) Magyar ügyszám: E 03 026690 (22) A bejelentés napja: 03.
Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092)
 Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092) www.zoolog.hu Dr. Dombos Miklós Tudományos főmunkatárs MTA ATK TAKI Innovative Real-time Monitoring and Pest control
Hasznos és kártevő rovarok monitorozása innovatív szenzorokkal (LIFE13 ENV/HU/001092) www.zoolog.hu Dr. Dombos Miklós Tudományos főmunkatárs MTA ATK TAKI Innovative Real-time Monitoring and Pest control
Supplementary Table 2. Summary of putative S-nitrosylated cysteines under nitrosative stress in C. reinhardtii.
 Supplementary Table 2. Summary of putative S-nitrosylated cysteines under nitrosative stress in C. reinhardtii. Accession number: accession number in UniprotKB protein database. RefSeq accession number:
Supplementary Table 2. Summary of putative S-nitrosylated cysteines under nitrosative stress in C. reinhardtii. Accession number: accession number in UniprotKB protein database. RefSeq accession number:
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon
 Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter
 Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter DOI: http://doi.org/10.13140/rg.2.2.28994.22721 A tudományos közlemények írása minden szakma művelésének
Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter DOI: http://doi.org/10.13140/rg.2.2.28994.22721 A tudományos közlemények írása minden szakma művelésének
A SZEMCSEALAK ALAPJÁN TÖRTÉNŐ SZÉTVÁLASZTÁS JELENTŐSÉGE FÉMTARTALMÚ HULLADÉKOK FELDOLGOZÁSA SORÁN
 Műszaki Földtudományi Közlemények, 83. kötet, 1. szám (2012), pp. 61 70. A SZEMCSEALAK ALAPJÁN TÖRTÉNŐ SZÉTVÁLASZTÁS JELENTŐSÉGE FÉMTARTALMÚ HULLADÉKOK FELDOLGOZÁSA SORÁN SIGNIFICANCE OF SHAPE SEPARATION
Műszaki Földtudományi Közlemények, 83. kötet, 1. szám (2012), pp. 61 70. A SZEMCSEALAK ALAPJÁN TÖRTÉNŐ SZÉTVÁLASZTÁS JELENTŐSÉGE FÉMTARTALMÚ HULLADÉKOK FELDOLGOZÁSA SORÁN SIGNIFICANCE OF SHAPE SEPARATION
DG(SANCO)/2012-6290-MR
 1 Ensure official controls of food contaminants across the whole food chain in order to monitor the compliance with the requirements of Regulation (EC) No 1881/2006 in all food establishments, including
1 Ensure official controls of food contaminants across the whole food chain in order to monitor the compliance with the requirements of Regulation (EC) No 1881/2006 in all food establishments, including
Azobezitás és a sejtek metabolizmusának összefüggései, a diabetes és táplálkozás viszonya
 Azobezitás és a sejtek metabolizmusának összefüggései, a diabetes és táplálkozás viszonya Máthé Endre, Sipos Péter, Remenyik Judit, Vígh Szabolcs, Horváth Brigitta Debreceni Egyetem, Mezőgazdaság, Élelmiszertudományi
Azobezitás és a sejtek metabolizmusának összefüggései, a diabetes és táplálkozás viszonya Máthé Endre, Sipos Péter, Remenyik Judit, Vígh Szabolcs, Horváth Brigitta Debreceni Egyetem, Mezőgazdaság, Élelmiszertudományi
Tavaszi Sporttábor / Spring Sports Camp. 2016. május 27 29. (péntek vasárnap) 27 29 May 2016 (Friday Sunday)
 Tavaszi Sporttábor / Spring Sports Camp 2016. május 27 29. (péntek vasárnap) 27 29 May 2016 (Friday Sunday) SZÁLLÁS / ACCOMODDATION on a Hotel Gellért*** szálloda 2 ágyas szobáiban, vagy 2x2 ágyas hostel
Tavaszi Sporttábor / Spring Sports Camp 2016. május 27 29. (péntek vasárnap) 27 29 May 2016 (Friday Sunday) SZÁLLÁS / ACCOMODDATION on a Hotel Gellért*** szálloda 2 ágyas szobáiban, vagy 2x2 ágyas hostel
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel
 Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
A teszt a következő diával indul! The test begins with the next slide!
 A teszt a következő diával indul! The test begins with the next slide! A KÖVETKEZŐKBEN SZÁMOZOTT KÉRDÉSEKET VAGY KÉPEKET LÁT SZÁMOZOTT KÉPLETEKKEL. ÍRJA A SZÁMOZOTT KÉRDÉSRE ADOTT VÁLASZT, VAGY A SZÁMOZOTT
A teszt a következő diával indul! The test begins with the next slide! A KÖVETKEZŐKBEN SZÁMOZOTT KÉRDÉSEKET VAGY KÉPEKET LÁT SZÁMOZOTT KÉPLETEKKEL. ÍRJA A SZÁMOZOTT KÉRDÉSRE ADOTT VÁLASZT, VAGY A SZÁMOZOTT
ÖNHORDÓ BORDÁZOTT LEMEZ
 ÖNHORDÓ BORDÁZOTT LEMEZ Az ilyen típusú profil fő alkalmazása - beton zsaluzat. Két változatban gyártják: egyszerű profil, vagy bordázott profil, hogy biztosítsa a beton és a profil közötti jobb együtthatást.
ÖNHORDÓ BORDÁZOTT LEMEZ Az ilyen típusú profil fő alkalmazása - beton zsaluzat. Két változatban gyártják: egyszerű profil, vagy bordázott profil, hogy biztosítsa a beton és a profil közötti jobb együtthatást.
Crash Course in Omics Terminology, Concepts & Data Types
 Crash Course in Omics Terminology, Concepts & Data Types 14 th Annual EuPathDB Workshop Jessie Kissinger June 2, 2019 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB GiardiaDB
Crash Course in Omics Terminology, Concepts & Data Types 14 th Annual EuPathDB Workshop Jessie Kissinger June 2, 2019 FungiDB MicrosporidiaDB AmoebaDB PlasmoDB ToxoDB CryptoDB PiroplasmaDB TrichDB GiardiaDB
PIAL1 (At1g08910) ORF sequence
 PIAL1 (At1g08910) ORF sequence ATGGTTATTCCGGCGACTTCTAGGTTTGGGTTTCGTGCTGAATTCAACACCAAGGAGTTTCAAGCTTCCTGCATCTCTCT! CGCTAATGAAATCGATGCGGCTATTGGGAGAAATGAAGTTCCAGGGAATATTCAAGAGCTCGCTTTAATCCTCAATAATG! TGTGCCGACGTAAATGTGATGATTATCAAACCAGGGCGGTGGTAATGGCGCTGATGATCTCAGTCAAGAGTGCTTGTCAG!
PIAL1 (At1g08910) ORF sequence ATGGTTATTCCGGCGACTTCTAGGTTTGGGTTTCGTGCTGAATTCAACACCAAGGAGTTTCAAGCTTCCTGCATCTCTCT! CGCTAATGAAATCGATGCGGCTATTGGGAGAAATGAAGTTCCAGGGAATATTCAAGAGCTCGCTTTAATCCTCAATAATG! TGTGCCGACGTAAATGTGATGATTATCAAACCAGGGCGGTGGTAATGGCGCTGATGATCTCAGTCAAGAGTGCTTGTCAG!
