Supplemental Information. Pervasive within-mitochondrion. Single-Nucleotide Variant Heteroplasmy. as Revealed by Single-Mitochondrion Sequencing
|
|
|
- János Csonka
- 5 évvel ezelőtt
- Látták:
Átírás
1 ell Reports, Volume 21 Supplemental Information Pervasive within-mitochondrion Single-Nucleotide Variant Heteroplasmy as Revealed by Single-Mitochondrion Sequencing Jacqueline Morris, Young-Ji Na, Hua Zhu, Jae-Hee Lee, Hoa iang, Alexandra V. Ulyanova, ordon H. Baltuch, Steven Brem, H. Isaac hen, David K. Kung, imothy H. Lucas, Donald M. O'Rourke, John A. Wolf, M. Sean rady, Jai-Yoon Sul, Junhyong Kim, and James Eberwine
2 Supplemental Materials: Inventory of Supplement: Supplemental Figure 1. Sample lineage tree depicting the relationship between samples. his figure shows the pedigree of all of the samples processed in this study, with Figure 2 in the main text highlighting important relationships between a subset of samples derived from the same pedigree (mother). Supplemental Figure 2. Sample lineage tree scaled to SNV events along each lineage. his figure shows the mutational burden in each sample in the context of the relationships between samples (pedigree), with Figure 2 in the main text highlighting pedigrees with unusual mutational burden compared to the others. Supplemental Figure 3. Relationship between mouse or human single mitochondrion samples and allele identities at high confidence positions. his figure shows the mutations present for mother and pup at the high confidence positions identified in both mouse and human, which are characterized further in Figure 2 and Figure 3 in the main text, respectively. Supplemental able 1. Primer sequences used for PR amplification of mouse single mitochondrion genomic samples. his table provides the sequences of primers used to achieve the amplification strategy highlighted in Figure 1. Supplemental able 2. Primer sequences used for PR amplification of human single mitochondrion genomic samples. his table provides the sequences of primers used to achieve the amplification strategy highlighted in Figure 1. Supplemental Information. Experimental Methods and PR error model for calling single nucleotide variants. Additional experimental methods used in this work are presented here. Additionally, this supplement provides more detailed mathematical descriptions of how a threshold was set for calling a mutation as a single nucleotide variant.
3 Fig. S1. Sample lineage tree depicting the relationship between samples. Related to Figure 2. Each sample is represented by the generic code MxxDx[N/A/][h/c/b] where x is a number. Mx = mother ID. x = cell ID. x is appended with a lowercase letter for samples isolated from the same cell. Dx = dendrite ID, Dx = 0 for astrocytes. [N/A/] = cell type, where N=neuron, A=astrocyte, =population. [h/c/b] = brain region from which the sample was derived: h=hippocampus, c=cortex, b=cerebellum. A reference population sample is grouped by itself on the left of the single mitochondrion samples
4 Fig. S2. Sample lineage tree scaled to SNV events along each lineage. Related to Figure 2. he branch lengths of the pictured cell lineage tree are scaled by estimated mutation events along each branch. he Maximum Parsimony method (see Methods) of character reconstruction was used to estimate for each branch the mutation events for the positions with changes in the major allele as well as those positions with changes in the minor allele.
5 A I I () I I A() I,() I (), I A() I,() I (), M27178aD0Ac M27178bD0Ac M27179D0Ac M27186D4Nc M27186D1Nc M27181D0Ac M27182bD0Ac M27182aD0Ac M27182cD0Ac M27184cD1Nc M27184bD1Nc M27184aD1Nc M27185D1Nc M27183aD1Nc M27183bD1Nc M29188D0Ac M29190D0Ac M29189D1Nc M29187D0Ac M30191aD0Ac M30191bD0Ac 3816() 9027() A (A) A() A() A 9461() () () () () () () () () () () () () 16265() () () () () () () () () () B M11P1N (S1) M11P1N (S2) M11P1N (S3) M11P1N (S4) M11P1N (S5) M13X (S6) M12P1N (S7) M12P1N (S8) M15P1N (S9) M15P1N (S10) M15P1N (S11) M15P1N (S12) M15P1N (S13) M15P1N (S14) M15P1N (S15) M15P1N (S16) M15P1N (S17) M16X (S18) M17X (S19) M19X (S20) M110X (S21) 309() () () () () () () () () () () () () () () () () () () () () () Fig. S3. Relationship between mouse or human single mitochondrion samples and allele identities at high confidence positions. Related to Figure 2 and Figure 3. Sample lineage trees of mouse (A) or human (B) single mitochondrion with Major(minor) alleles indicated for each high confidence position. he reference allele is indicated in parentheses next to each position label. Major(minor) alleles are listed for mouse mother and pup at the beginning of each branch point for each mother for each position. here is a single entry when the mother and pup alleles agreed. Human sample code format is MxxPx where M =mother ID, = cell ID, P = process ID, =cell type where N = neuronal-like or X = non-neuronal, with simplified codes in parentheses.
6 able S1. Primer sequences used for PR amplification of mouse single mitochondrion genomic samples. Related to Figure 1. Version 1 Long PR Primers Forward Reverse Amplicon 1 AAAAAAAA AAAAA Amplicon 2 AAAAA AAAAA Amplicon 3 AAA AAAA Version 2 Long PR Primers Forward Reverse Amplicon 1 AAAAAAA AAAAAA Amplicon 2 AAAAA AAAA Amplicon 3 AAAAAAA AA Short PR primers Forward Reverse Set 1 AAAAAAAA AAA Set 2 AAAAA AA Set 3 AA AAAAA Set 4 AAAAA AAAAAAAAA Set 5 AAAA AAAAAAAAAA Set 6 AAAAA AAAAA Set 7 AAA AAA Set 8 AAAAA AAA Set 9 AAA AAAA Set 10 AAAAAAAA AA Set 11 AAAAAAA AA Set 12 AAAAAA AAA Set 13 AAAAAAA AAAAA Set 14 AAAAA AAAAA Set 15 AAA AA Set 16 AAAAAAAAAA AAAA Set 17 AAA AAAAA Set 18 AAA AA Set 19 AAAAAA AA Set 20 AAAAAAAAA AAAAA Set 21 AAAA AAAA Set 22 AAAAAAAA AA Set 23 AAAAA AAAAAAA Set 24 AAAAAA AAAAA Set 25 AAAAA AAAAAA Set 26 AAAAA AAAAA Set 27 AAAAAA AAAAA Set 28 AAA AAAA Set 29 AAAAAAA AAAAA Set 30 AAAAA AAAA Set 31 AAAAAAA AAAAA Set 32 AAAAAAA AAAAA Set 33 AAAAA AAAAAA Set 34 AAAAA AAAAAAA Set 35 AAAAA AAAA Set 36 AAAAA AAAA Set 37 AAAAAAA AAAAAA Set 38 AAAAAAAAA AAAA Set 39 AAAAA AA Set 40 AAAAAAA AAA Set 41 AAAAAA AAAA Set 42 AAAAA AAAAAA Set 43 AAAAA AAAAAAA Set 44 AAAA AAAAAAA Set 45 AAAAA AAAA Set 46 AAAAAAA AAA Set 47 AAA AAAAAA Set 48 AAAAAA AA Set 49 AAAAAAA AAAA Set 50 AAA AAAA Set 51 AAAAA AAAAAA Set 52 AAAAA AAAA Set 53 AAAAAA AAAA Set 54 AAAAAAA AAAA Set 55 AAAAAAA AAAAA Set 56 AAAAAA AA Forward and reverse primer sequences are listed for both the long PR (Round 1 PR) and short PR (Round 2 PR) reactions.
7 able S2. Primer sequences used for PR amplification of human single mitochondrion genomic samples. Related to Figure 1. Long PR Primers Forward Reverse Amplicon 1 AAAAA AAAA Amplicon 2 AAAAAAAA AAAAAA Amplicon 3 AAAAAA AAAAAAA Short PR primers Forward Reverse Set 1 AAAAA AA Set 2 AAAAAA AAAA Set 3 AAAA AAAA Set 4 AAAAAAAA AAAAAAAAA Set 5 AAAAA AAAA Set 6 AAAAA AAAAAA Set 7 AAAAAA AAAAAA Set 8 AAAAA AAAAA Set 9 AAAAAAAA AAAAAAA Set 10 AAAAA AAAAAA Set 11 AAAAAA AAAA Set 12 AAAAA AAAA Set 13 AAAAA AAAA Set 14 AAAAAAAA AAAA Set 15 AAAAA AAAA Set 16 AAAAA AAA Set 17 AAAA AAAAAA Set 18 AAAAAA A Set 19 AAAA AAAA Set 20 AAAAAA AAAAAAA Set 21 AAAAA AAAA Set 22 AAAAAA AA Set 23 AAA AAAA Set 24 AAAAAAAA AAAAA Set 25 AAAAAAA AA Set 26 AAAA AAAAAA Set 27 AAAAAA AA Set 28 AAAAAA AA Set 29 AAAAA AAAAAAA Forward and reverse primer sequences are listed for both the long PR (Round1 PR) and short PR (Round 2 PR) reactions.
8 Supplemental Experimental Procedures: Subjects. Mouse tissue was collected under University of Pennsylvania IAU Protocol# Postnatal day 1 pups from which cultures were derived were not sexed for these studies. Pups were not sexed because cultures were derived from multiple pups provided by a shared resource. Human brain tissue was collected at the Hospital of the University of Pennsylvania under Institutional Review Board approval # , using standard operating procedures for enrollment and consent of patients for patient numbers 50 (63 year old female) and 8 (65 year old female). See Supplemental Materials. ell ulture. Primary cortical neuronal, astrocyte or combination cultures were plated and maintained as described previously with some modifications (Kaech and Banker, 2006). Briefly, cortical neurons and/or astrocytes were isolated from postnatal day 1 mouse pups and dissociated with trypsin followed by pipetting prior to live cell counting with trypan blue and plating on poly-d-lysine/laminin coated 12-mm coverslips. Neuronal cultures were maintained in MEM with B27 supplement, astrocyte cultures in MEM with 10% FBS, co-cultures in MEM with B27 supplement and 1% FBS. ultures of human cells were prepared and maintained as described previously (Spaethling et al., 2017). A mm block of cortical tissue was resected as part of a neurosurgical procedure for the treatment of epilepsy or brain tumors. For transport (approximately 10 minutes from surgery to lab), this tissue was immediately transferred to a sterile tube containing ice-cold asf (2 mm al 2-2H 2 O, 10 mm glucose, 3 mm Kl, 26 mm NaHO 3, 2.5 mm NaH 2 PO 4, 1 mm Mgl 2-6H 2 O, and 202 mm sucrose, perfused with 95%/5% O 2 /O 2 ). Upon receipt, tissue was digested with papain (20 U, Worthington Biochemical) and incubated for min at 37. he reaction was stopped by addition of Leupeptin (a papain inhibitor, 100 µm, Sigma-Aldrich. Dissociated tissue was centrifuged (1,500 rpm for 3 min) and the supernatant subjected to gentle mechanical dissociation with a fire-polished glass Pasteur pipette. Live cells were stained with trypan blue and counted using an Autocounter (Invitrogen). ells were plated on poly-l-lysine-coated 12-mm coverslips at a density of cells/coverslip. ultures were maintained at 37 under 95% humidity and 5% O 2 in medium (Neurobasal supplemented with B27 (1%) and penicillin/streptomycin (1%). he medium was changed by replacing 50% with fresh medium every 3 days. Primer Design. All primers were designed by exhaustive computational analysis to minimize secondary structure and cross interactions and to maintain compatible melting temperatures within each set of PR primers. he nested set of long PR primers were designed by first generating three sets of primers that would create amplicons of approximately 5 Kb such that each set was offset from each other. We then designed short PR primers again with overlapping offsets such that the set of amplicons exhaustively covered the long PR amplicons (Fig. 1B). Mitotracker Loading and imaging of cells. One coverslip of cells was transferred to one well of a 4 well dish containing 25 nm Mitotracker Red MXRos (hermofisher Scientific, M-7512) in 500 µl 50% conditioned/50% fresh media. Mitotracker loading was performed at 37 with 5% O 2 for 20 minutes before transferring the coverslip to fresh 50% conditioned/50% fresh media and returning to the incubator for 10 minutes. he coverslip was then transferred to a low profile imaging chamber for imaging and sample collection. Samples were imaged with a Zeiss 710 LSM using the Alexa568 preset filter settings to image Mitotracker Red MXRos and transmitted light for cell morphology. Samples were continuously perfused with normal saline (140 mm Nal, 5.4 mm Kl, 1 mm Mgl 2, 2 mm al 2, 16 mm glucose, 10 mm HEPES, ph 7.3) in between isolations. Mitochondrial DNA Library construction and Sequencing. Library construction was performed using the Ilumina ruseq DNA nano library construction kit according to manufacturer s instructions. Successful short PR reactions were pooled together and cleaned with a 0.5x AMPURE XP bead clean up. Based on concentrations of the pooled short PR reactions obtained from running samples on an Agilent DNA 7500 chip on the Agilent Bioanalyzer, 247 ng of each pool was made up in a final volume of 130 µl of resuspension buffer, transferred to an AFA Fiber Pre-Slit Snap-ap 6x16 mm microube (ovaris ) and sheared on a ovaris S2 focusedultrasonicator with the following settings to achieve an average insert size of 350 bp: Duty ycle:10%, Intensity:5, ycles/burst:200, ime: 45s). Sheared DNA was then quantified on an Agilent High Sensitivity DNA kit on the Agilent Bioanalyzer or the Agilent High Sensitivity D5000 kit on the Agilent apestation 2200 before inputting into the 350 bp insert workstream of the library kit protocol. Single mitochondrion libraries were pooled 24 samples per lane and run in rapid mode on a HiSeq 2500 with 2x101bp paired end reads. Variant calling from RNASeq. he SNPiR pipeline (Piskol et al., 2013) was implemented as follows: he burrows-wheeler aligner (BWA) was used to map RNA-Seq reads derived from sequencing of mouse or human single cells using the commands bwa aln fastqfile and bwa samse n4. We only considered positions covered by the RNA-Seq for variant calling if the following criteria were met: (A) read depth coverage > 50, (B) basequality >
9 0, () mappingquality > 0, (D) read frequency of minor allele 0.04%. SAMtools rmdup (Li et al., 2009) was used to remove identical reads derived from PR duplicates that mapped to the same location. Effect of variants on trna and protein structure. For position 3816, located in the gene encoding the mitochondrial trna for glutamine, the secondary structure was obtained from the mitotrnadb maintained by the Universities of Leipzig and Strasbourg and the output structure used to indicate the location of the variable position in this trna (Jühling et al., 2009). For position 9027, located in the gene encoding mitochondrial cytochrome c oxidase subunit 3, the secondary and tertiary structure of this subunit was modeled by inputting UnitProt entry number P00416 (OX3_MOUSE) into the protein model portal for structural analysis (Schwede et al., 2009). he structure was inferred from the 85% sequence homology with bovine (Bos aurus) heart cytochrome c oxidase protein in the fully oxidized state (1V54), whose structure was determined by x-ray crystallography at a resolution of 1.8 angstroms. At this level of structure resolution and sequence identity, the structural predications for the target (here the mouse protein) are typically correct. his particular subunit of the OX enzyme is highly conserved across Eukaryotes based on protein sequence alignments. PR error model. Here we use a model-based calculation to derive the probability of observing variant allele whose read frequency is greater than some threshold value, θ. Assume N cycles of PR reactions resulting in 2 N total molecules. If a mutation occurs in the 1 st PR cycle, then 50% of the final molecules will have this mutation. In general, if a mutation happens in the ith cycle,!!!! =!! 2!! proportion of the final molecules will have this mutation. Let n be the average sequencing depth, then we assume that out of 2 N total final molecules (including the library prep PR cycles), we have sampled n total molecules. Let X be the number of sequenced reads carrying a non-reference mutation. We wish to compute P X > nθ n, i. Let P(i) denote the probability of a PR mutation in the ith cycle. hen P X > nθ n =!!!! P X > nθ n, i P(i) (1) he probability P(i) of at least one mutation in the ith cycle is P(i) = 1 (1 ε)!!!! (2) where ε is the PR polymerase mutation rate. We note that (1) ignores the O(ε! ) probability of convergent multiple mutations in the ith cycle. he term!! =! 2!!! denotes the number of lineages in the ith cycle where mutations may occur, where ½ is due to semi-conservative replication. aq polymerase error rates are estimated to be between 1 10!! to 1 10!!. herefore, using (2) for aq, P(i)\ycle Min 1 10!! 2 10!! 4 10!! 8 10!! Max 1 10!! 2 10!! 4 10!! 8 10!! he probability of sampling a molecule carrying a mutation from the ith cycle is 2!! and we use the normal approximation to the binomial sampling process and assume the number X of sequenced molecules with variant mutation is X~N(n 2!!, n 2!! 1 2!! ). In our experiments, our average sequencing depth was ~17,500; or, the number of molecules sampled was n = 17,500. Under these parameters P X > nθ n, i is approximately 0 for values of i > 3. For values of i 3, P X > nθ n, i is either approximately 1 or 0 for various values of θ. able below shows the values of P X > nθ n, i for θ = 0.1, 0.2, 0.3, 0.4, and 0.5. ycle i\θ ~1 ~1 ~1 ~1 0.5
10 2 ~1 ~1 ~0 ~0 ~0 3 ~1 ~0 ~0 ~0 ~0 4 ~0 ~0 ~0 ~0 ~0 ombining the two tables in Equation (1), we obtain the following table for P(X > nθ n = 17,500): able 1: aq-related probability of observing false positive variant at frequency greater than θ. P X > nθ n, i \θ Min 7 10!! 3 10!! 1 10!! 1 10!! !! Max 7 10!! 3 10!! 1 10!! 1 10!! !! For the enzyme Q5, the estimates of error rates are more than 100-fold lower. If Q5 enzyme is used in the 1 st four PR cycles, able 1 can be simply scaled to able 2: Q5-related probability of observing false positive variant at frequency greater than θ. P X > nθ n, i \θ Min 7 10!! 3 10!! 1 10!! 1 10!! !! Max 7 10!! 3 10!! 1 10!! 1 10!! !! Shared SNV sites for multiple mitochondria. Based on the PR-derived false positive rate per site, the probability of observing k or more samples with a variant call at a particular site by misincorporation error is approximately n, k !!!, where n =118 is the number of samples in our mouse dataset. Using conservative Bonferroni correction over the ~16,000 sites of the mitochondrial genome, the probability, p, of observing k = 2, 3, 4 samples with a variant call are, p > 1, 0.5, , respectively. For human samples with n =22, the Bonferroni corrected probability of k = 2, 3, 4 samples are p = , , and > 10-7, respectively. Supplemental References: Kaech, S., Banker,. (2006). ulturing hippocampal neurons. Nat. Protoc. 1, Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, Piskol, R., Ramaswami,., Li, J.B. (2013). Reliable identification of genomic variants from RNA-Seq data. Am. J. Hum. enet. 93, Jühling, F., Mörl, M., Hartmann, R.K., Sprinzl, M., Stadler, P.F., Pütz, J. (2009). trnadb 2009: compilation of trna sequences and trna genes. Nucleic Acids Res. 37, D159-D162. Schwede,., Sali, A., Honig, B., Levitt, M., Berman, H.M., Jones, D., Brenner, S.E., Burley, S.K., Das, R., Dokholyan, N.V. et al. (2009). Outcome of a Workshop on Applications of Protein Models in Biomedical Research. Structure 17,
Mapping Sequencing Reads to a Reference Genome
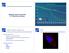 Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Supporting Information
 Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Supplementary Figure 1
 Supplementary Figure 1 Plot of delta-afe of sequence variants detected between resistant and susceptible pool over the genome sequence of the WB42 line of B. vulgaris ssp. maritima. The delta-afe values
Supplementary Figure 1 Plot of delta-afe of sequence variants detected between resistant and susceptible pool over the genome sequence of the WB42 line of B. vulgaris ssp. maritima. The delta-afe values
Supplementary Table 1. Cystometric parameters in sham-operated wild type and Trpv4 -/- rats during saline infusion and
 WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo
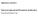 Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
SOLiD Technology. library preparation & Sequencing Chemistry (sequencing by ligation!) Imaging and analysis. Application specific sample preparation
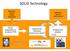 SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing MFMER slide-1
 Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
University of Bristol - Explore Bristol Research
 Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Correlation & Linear Regression in SPSS
 Correlation & Linear Regression in SPSS Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise 1 - Correlation File / Open
Correlation & Linear Regression in SPSS Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise 1 - Correlation File / Open
Correlation & Linear Regression in SPSS
 Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
Petra Petrovics Correlation & Linear Regression in SPSS 4 th seminar Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation
Statistical Inference
 Petra Petrovics Statistical Inference 1 st lecture Descriptive Statistics Inferential - it is concerned only with collecting and describing data Population - it is used when tentative conclusions about
Petra Petrovics Statistical Inference 1 st lecture Descriptive Statistics Inferential - it is concerned only with collecting and describing data Population - it is used when tentative conclusions about
Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes
 Stem Cell Reports, Volume 2 Supplemental Information Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes Yuko
Stem Cell Reports, Volume 2 Supplemental Information Involvement of ER Stress in Dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 Missense Mutations Shown by ipsc-derived Oligodendrocytes Yuko
Forensic SNP Genotyping using Nanopore MinION Sequencing
 Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Correlation & Linear. Petra Petrovics.
 Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
Correlation & Linear Regression in SPSS Petra Petrovics PhD Student Types of dependence association between two nominal data mixed between a nominal and a ratio data correlation among ratio data Exercise
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet Nonparametric Tests
 Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Genome 373: Hidden Markov Models I. Doug Fowler
 Genome 373: Hidden Markov Models I Doug Fowler Review From Gene Prediction I transcriptional start site G open reading frame transcriptional termination site promoter 5 untranslated region 3 untranslated
Genome 373: Hidden Markov Models I Doug Fowler Review From Gene Prediction I transcriptional start site G open reading frame transcriptional termination site promoter 5 untranslated region 3 untranslated
FAMILY STRUCTURES THROUGH THE LIFE CYCLE
 FAMILY STRUCTURES THROUGH THE LIFE CYCLE István Harcsa Judit Monostori A magyar társadalom 2012-ben: trendek és perspektívák EU összehasonlításban Budapest, 2012 november 22-23 Introduction Factors which
FAMILY STRUCTURES THROUGH THE LIFE CYCLE István Harcsa Judit Monostori A magyar társadalom 2012-ben: trendek és perspektívák EU összehasonlításban Budapest, 2012 november 22-23 Introduction Factors which
discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo
 discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo Download and install discosnp demo - Peterlongo Pierre 3 Download web page: github.com/gatb/discosnp Chose latest release (2.2.10 today) discosnp
discosnp demo - Peterlongo Pierre 1 DISCOSNP++: Live demo Download and install discosnp demo - Peterlongo Pierre 3 Download web page: github.com/gatb/discosnp Chose latest release (2.2.10 today) discosnp
Expression analysis of PIN genes in root tips and nodules of Lotus japonicus
 Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Széchenyi István Egyetem www.sze.hu/~herno
 Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
EN United in diversity EN A8-0206/419. Amendment
 22.3.2019 A8-0206/419 419 Article 2 paragraph 4 point a point i (i) the identity of the road transport operator; (i) the identity of the road transport operator by means of its intra-community tax identification
22.3.2019 A8-0206/419 419 Article 2 paragraph 4 point a point i (i) the identity of the road transport operator; (i) the identity of the road transport operator by means of its intra-community tax identification
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Hypothesis Testing. Petra Petrovics.
 Hypothesis Testing Petra Petrovics PhD Student Inference from the Sample to the Population Estimation Hypothesis Testing Estimation: how can we determine the value of an unknown parameter of a population
Hypothesis Testing Petra Petrovics PhD Student Inference from the Sample to the Population Estimation Hypothesis Testing Estimation: how can we determine the value of an unknown parameter of a population
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Nonparametric Tests. Petra Petrovics.
 Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
Statistical Dependence
 Statistical Dependence Petra Petrovics Statistical Dependence Deinition: Statistical dependence exists when the value o some variable is dependent upon or aected by the value o some other variable. Independent
Statistical Dependence Petra Petrovics Statistical Dependence Deinition: Statistical dependence exists when the value o some variable is dependent upon or aected by the value o some other variable. Independent
A cell-based screening system for RNA Polymerase I inhibitors
 Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet Factor Analysis
 Factor Analysis Factor analysis is a multiple statistical method, which analyzes the correlation relation between data, and it is for data reduction, dimension reduction and to explore the structure. Aim
Factor Analysis Factor analysis is a multiple statistical method, which analyzes the correlation relation between data, and it is for data reduction, dimension reduction and to explore the structure. Aim
Supporting Information
 Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Construction of a cube given with its centre and a sideline
 Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma
 T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma Prescott G. Woodruff, M.D., M.P.H., Barmak Modrek, Ph.D., David F. Choy, B.S., Guiquan Jia, M.D., Alexander R. Abbas, Ph.D., Almut
T-helper Type 2 driven Inflammation Defines Major Subphenotypes of Asthma Prescott G. Woodruff, M.D., M.P.H., Barmak Modrek, Ph.D., David F. Choy, B.S., Guiquan Jia, M.D., Alexander R. Abbas, Ph.D., Almut
A rosszindulatú daganatos halálozás változása 1975 és 2001 között Magyarországon
 A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
A rosszindulatú daganatos halálozás változása és között Eredeti közlemény Gaudi István 1,2, Kásler Miklós 2 1 MTA Számítástechnikai és Automatizálási Kutató Intézete, Budapest 2 Országos Onkológiai Intézet,
FÖLDRAJZ ANGOL NYELVEN
 Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
Földrajz angol nyelven középszint 0821 ÉRETTSÉGI VIZSGA 2009. május 14. FÖLDRAJZ ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI ÉRETTSÉGI VIZSGA JAVÍTÁSI-ÉRTÉKELÉSI ÚTMUTATÓ OKTATÁSI ÉS KULTURÁLIS MINISZTÉRIUM Paper
STUDENT LOGBOOK. 1 week general practice course for the 6 th year medical students SEMMELWEIS EGYETEM. Name of the student:
 STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
STUDENT LOGBOOK 1 week general practice course for the 6 th year medical students Name of the student: Dates of the practice course: Name of the tutor: Address of the family practice: Tel: Please read
On The Number Of Slim Semimodular Lattices
 On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley M. Lemon, Lawrence M. Pfeffer, Kui Li
 Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
16F628A megszakítás kezelése
 16F628A megszakítás kezelése A 'megszakítás' azt jelenti, hogy a program normális, szekvenciális futása valamilyen külső hatás miatt átmenetileg felfüggesztődik, és a vezérlést egy külön rutin, a megszakításkezelő
16F628A megszakítás kezelése A 'megszakítás' azt jelenti, hogy a program normális, szekvenciális futása valamilyen külső hatás miatt átmenetileg felfüggesztődik, és a vezérlést egy külön rutin, a megszakításkezelő
Modular Optimization of Hemicellulose-utilizing Pathway in. Corynebacterium glutamicum for Consolidated Bioprocessing of
 [Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
[Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
Limitations and challenges of genetic barcode quantification
 Limitations and challenges of genetic barcode quantification Lars hielecke, im ranyossy, ndreas Dahl, Rajiv iwari, Ingo Roeder, Hartmut eiger, Boris Fehse, Ingmar lauche and Kerstin ornils SUPPLEMENRY
Limitations and challenges of genetic barcode quantification Lars hielecke, im ranyossy, ndreas Dahl, Rajiv iwari, Ingo Roeder, Hartmut eiger, Boris Fehse, Ingmar lauche and Kerstin ornils SUPPLEMENRY
Phenotype. Genotype. It is like any other experiment! What is a bioinformatics experiment? Remember the Goal. Infectious Disease Paradigm
 It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
PIACI HIRDETMÉNY / MARKET NOTICE
 PIACI HIRDETMÉNY / MARKET NOTICE HUPX DAM Másnapi Aukció / HUPX DAM Day-Ahead Auction Iktatási szám / Notice #: Dátum / Of: 18/11/2014 HUPX-MN-DAM-2014-0023 Tárgy / Subject: Változások a HUPX másnapi piac
PIACI HIRDETMÉNY / MARKET NOTICE HUPX DAM Másnapi Aukció / HUPX DAM Day-Ahead Auction Iktatási szám / Notice #: Dátum / Of: 18/11/2014 HUPX-MN-DAM-2014-0023 Tárgy / Subject: Változások a HUPX másnapi piac
Tudományos Ismeretterjesztő Társulat
 Sample letter number 5. International Culture Festival PO Box 34467 Harrogate HG 45 67F Sonnenbergstraße 11a CH-6005 Luzern Re: Festival May 19, 2009 Dear Ms Atkinson, We are two students from Switzerland
Sample letter number 5. International Culture Festival PO Box 34467 Harrogate HG 45 67F Sonnenbergstraße 11a CH-6005 Luzern Re: Festival May 19, 2009 Dear Ms Atkinson, We are two students from Switzerland
Flowering time. Col C24 Cvi C24xCol C24xCvi ColxCvi
 Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Márkaépítés a YouTube-on
 Márkaépítés a YouTube-on Tv+ Adj hozzá YouTube-ot, Google Ground, 2016 Március 7. Bíró Pál, Google - YouTube 9,000,000 INTERNETTEL BÍRÓ ESZKÖZÖK VOLUMENE GLOBÁLISAN WEARABLES OKOS TV 8,000,000 7,000,000
Márkaépítés a YouTube-on Tv+ Adj hozzá YouTube-ot, Google Ground, 2016 Március 7. Bíró Pál, Google - YouTube 9,000,000 INTERNETTEL BÍRÓ ESZKÖZÖK VOLUMENE GLOBÁLISAN WEARABLES OKOS TV 8,000,000 7,000,000
CLUSTALW Multiple Sequence Alignment
 Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
Version 3.2 CLUSTALW Multiple Sequence Alignment Selected Sequences) FETA_GORGO FETA_HORSE FETA_HUMAN FETA_MOUSE FETA_PANTR FETA_RAT Import Alignments) Return Help Report Bugs Fasta label *) Workbench
Cashback 2015 Deposit Promotion teljes szabályzat
 Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
Gottsegen National Institute of Cardiology. Prof. A. JÁNOSI
 Myocardial Infarction Registry Pilot Study Hungarian Myocardial Infarction Register Gottsegen National Institute of Cardiology Prof. A. JÁNOSI A https://ir.kardio.hu A Web based study with quality assurance
Myocardial Infarction Registry Pilot Study Hungarian Myocardial Infarction Register Gottsegen National Institute of Cardiology Prof. A. JÁNOSI A https://ir.kardio.hu A Web based study with quality assurance
4 vana, vanb, vanc1, vanc2
 77 1) 2) 2) 3) 4) 5) 1) 2) 3) 4) 5) 16 12 7 17 4 8 2003 1 2004 7 3 Enterococcus 5 Enterococcus faecalis 1 Enterococcus avium 1 Enterococcus faecium 2 5 PCR E. faecium 1 E-test MIC 256 mg/ml vana MRSA MRSA
77 1) 2) 2) 3) 4) 5) 1) 2) 3) 4) 5) 16 12 7 17 4 8 2003 1 2004 7 3 Enterococcus 5 Enterococcus faecalis 1 Enterococcus avium 1 Enterococcus faecium 2 5 PCR E. faecium 1 E-test MIC 256 mg/ml vana MRSA MRSA
ENROLLMENT FORM / BEIRATKOZÁSI ADATLAP
 ENROLLMENT FORM / BEIRATKOZÁSI ADATLAP CHILD S DATA / GYERMEK ADATAI PLEASE FILL IN THIS INFORMATION WITH DATA BASED ON OFFICIAL DOCUMENTS / KÉRJÜK, TÖLTSE KI A HIVATALOS DOKUMENTUMOKBAN SZEREPLŐ ADATOK
ENROLLMENT FORM / BEIRATKOZÁSI ADATLAP CHILD S DATA / GYERMEK ADATAI PLEASE FILL IN THIS INFORMATION WITH DATA BASED ON OFFICIAL DOCUMENTS / KÉRJÜK, TÖLTSE KI A HIVATALOS DOKUMENTUMOKBAN SZEREPLŐ ADATOK
INDEXSTRUKTÚRÁK III.
 2MU05_Bitmap.pdf camü_ea INDEXSTRUKTÚRÁK III. Molina-Ullman-Widom: Adatbázisrendszerek megvalósítása Panem, 2001könyv 5.4. Bittérkép indexek fejezete alapján Oracle: Indexek a gyakorlatban Oracle Database
2MU05_Bitmap.pdf camü_ea INDEXSTRUKTÚRÁK III. Molina-Ullman-Widom: Adatbázisrendszerek megvalósítása Panem, 2001könyv 5.4. Bittérkép indexek fejezete alapján Oracle: Indexek a gyakorlatban Oracle Database
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1358 0.2.2 Species name Mustela putorius 0.2.3 Alternative species scientific name 0.2.4 Common name házigörény 1. National Level 1.1 Maps 1.1.1 Distribution Map
0.1 Member State HU 0.2.1 Species code 1358 0.2.2 Species name Mustela putorius 0.2.3 Alternative species scientific name 0.2.4 Common name házigörény 1. National Level 1.1 Maps 1.1.1 Distribution Map
Supporting Information
 Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
(NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV
 Kommunikációs rendszerek programozása (NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV (5. mérés) SIP telefonközpont készítése Trixbox-szal 1 Mérés helye: Széchenyi István Egyetem, L-1/7 laboratórium, 9026 Győr, Egyetem
Kommunikációs rendszerek programozása (NGB_TA024_1) MÉRÉSI JEGYZŐKÖNYV (5. mérés) SIP telefonközpont készítése Trixbox-szal 1 Mérés helye: Széchenyi István Egyetem, L-1/7 laboratórium, 9026 Győr, Egyetem
Proxer 7 Manager szoftver felhasználói leírás
 Proxer 7 Manager szoftver felhasználói leírás A program az induláskor elkezdi keresni az eszközöket. Ha van olyan eszköz, amely virtuális billentyűzetként van beállítva, akkor azokat is kijelzi. Azokkal
Proxer 7 Manager szoftver felhasználói leírás A program az induláskor elkezdi keresni az eszközöket. Ha van olyan eszköz, amely virtuális billentyűzetként van beállítva, akkor azokat is kijelzi. Azokkal
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Correlation & Regression
 Correlation & Regression Types of dependence association between nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation describes the strength of a relationship,
Correlation & Regression Types of dependence association between nominal data mixed between a nominal and a ratio data correlation among ratio data Correlation describes the strength of a relationship,
Rezgésdiagnosztika. Diagnosztika 02 --- 1
 Rezgésdiagnosztika Diagnosztika 02 --- 1 Diagnosztika 02 --- 2 A rezgéskép elemzésével kimutatható gépészeti problémák Minden gép, mely tartalmaz forgó részt (pl. motor, generátor, szivattyú, ventilátor,
Rezgésdiagnosztika Diagnosztika 02 --- 1 Diagnosztika 02 --- 2 A rezgéskép elemzésével kimutatható gépészeti problémák Minden gép, mely tartalmaz forgó részt (pl. motor, generátor, szivattyú, ventilátor,
The role of aristolochene synthase in diphosphate activation
 Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Mark Auspitz, Fayez Quereshy, Allan Okrainec, Alvina Tse, Sanjeev Sockalingam, Michelle Cleghorn, Timothy Jackson
 Mark Auspitz, Fayez Quereshy, Allan Okrainec, Alvina Tse, Sanjeev Sockalingam, Michelle Cleghorn, Timothy Jackson Division of General Surgery, University Health Network, University of Toronto, Toronto,
Mark Auspitz, Fayez Quereshy, Allan Okrainec, Alvina Tse, Sanjeev Sockalingam, Michelle Cleghorn, Timothy Jackson Division of General Surgery, University Health Network, University of Toronto, Toronto,
University of Bristol - Explore Bristol Research
 Tan, B. G., Wellesley, F. C., Savery, N. J., & Szczelkun, M. D. (2016). Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity.
Tan, B. G., Wellesley, F. C., Savery, N. J., & Szczelkun, M. D. (2016). Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity.
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 2633 0.2.2 Species name Mustela eversmanii 0.2.3 Alternative species scientific name 0.2.4 Common name molnárgörény 1. National Level 1.1 Maps 1.1.1 Distribution
0.1 Member State HU 0.2.1 Species code 2633 0.2.2 Species name Mustela eversmanii 0.2.3 Alternative species scientific name 0.2.4 Common name molnárgörény 1. National Level 1.1 Maps 1.1.1 Distribution
T Á J É K O Z T A T Ó. A 1108INT számú nyomtatvány a http://www.nav.gov.hu webcímen a Letöltések Nyomtatványkitöltő programok fülön érhető el.
 T Á J É K O Z T A T Ó A 1108INT számú nyomtatvány a http://www.nav.gov.hu webcímen a Letöltések Nyomtatványkitöltő programok fülön érhető el. A Nyomtatványkitöltő programok fület választva a megjelenő
T Á J É K O Z T A T Ó A 1108INT számú nyomtatvány a http://www.nav.gov.hu webcímen a Letöltések Nyomtatványkitöltő programok fülön érhető el. A Nyomtatványkitöltő programok fület választva a megjelenő
Személyes adatváltoztatási formanyomtatvány- Magyarország / Personal Data Change Form - Hungary
 Személyes adatváltoztatási formanyomtatvány- Magyarország / Personal Data Change Form - Hungary KITÖLTÉSI ÚTMUTATÓ: A formanyomtatványon a munkavállaló a személyes adatainak módosítását kezdeményezheti.
Személyes adatváltoztatási formanyomtatvány- Magyarország / Personal Data Change Form - Hungary KITÖLTÉSI ÚTMUTATÓ: A formanyomtatványon a munkavállaló a személyes adatainak módosítását kezdeményezheti.
Dr. Sasvári Péter Egyetemi docens
 A KKV-k Informatikai Infrastruktúrájának vizsgálata a Visegrádi országokban The Analysis Of The IT Infrastructure Among SMEs In The Visegrád Group Of Countries Dr. Sasvári Péter Egyetemi docens MultiScience
A KKV-k Informatikai Infrastruktúrájának vizsgálata a Visegrádi országokban The Analysis Of The IT Infrastructure Among SMEs In The Visegrád Group Of Countries Dr. Sasvári Péter Egyetemi docens MultiScience
Cluster Analysis. Potyó László
 Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Bird species status and trends reporting format for the period (Annex 2)
 1. Species Information 1.1 Member State Hungary 1.2.2 Natura 2000 code A634-B 1.3 Species name Ardea purpurea purpurea 1.3.1 Sub-specific population East Europe, Black Sea & Mediterranean/Sub-Saharan Africa
1. Species Information 1.1 Member State Hungary 1.2.2 Natura 2000 code A634-B 1.3 Species name Ardea purpurea purpurea 1.3.1 Sub-specific population East Europe, Black Sea & Mediterranean/Sub-Saharan Africa
A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke
 A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke A dokumentum A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásában
A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásának változási és hibajegyzéke A dokumentum A vitorlázás versenyszabályai a 2013-2016. évekre angol-magyar nyelvű kiadásában
Performance Modeling of Intelligent Car Parking Systems
 Performance Modeling of Intelligent Car Parking Systems Károly Farkas Gábor Horváth András Mészáros Miklós Telek Technical University of Budapest, Hungary EPEW 2014, Florence, Italy Outline Intelligent
Performance Modeling of Intelligent Car Parking Systems Károly Farkas Gábor Horváth András Mészáros Miklós Telek Technical University of Budapest, Hungary EPEW 2014, Florence, Italy Outline Intelligent
Bioinformatics: Blending. Biology and Computer Science
 Bioinformatics: Blending Biology and Computer Science MDNMSITNTPTSNDACLSIVHSLMCHRQ GGESETFAKRAIESLVKKLKEKKDELDSL ITAITTNGAHPSKCVTIQRTLDGRLQVAG RKGFPHVIYARLWRWPDLHKNELKHVK YCQYAFDLKCDSVCVNPYHYERVVSPGI DLSGLTLQSNAPSSMMVKDEYVHDFEG
Bioinformatics: Blending Biology and Computer Science MDNMSITNTPTSNDACLSIVHSLMCHRQ GGESETFAKRAIESLVKKLKEKKDELDSL ITAITTNGAHPSKCVTIQRTLDGRLQVAG RKGFPHVIYARLWRWPDLHKNELKHVK YCQYAFDLKCDSVCVNPYHYERVVSPGI DLSGLTLQSNAPSSMMVKDEYVHDFEG
A BÜKKI KARSZTVÍZSZINT ÉSZLELŐ RENDSZER KERETÉBEN GYŰJTÖTT HIDROMETEOROLÓGIAI ADATOK ELEMZÉSE
 KARSZTFEJLŐDÉS XIX. Szombathely, 2014. pp. 137-146. A BÜKKI KARSZTVÍZSZINT ÉSZLELŐ RENDSZER KERETÉBEN GYŰJTÖTT HIDROMETEOROLÓGIAI ADATOK ELEMZÉSE ANALYSIS OF HYDROMETEOROLIGYCAL DATA OF BÜKK WATER LEVEL
KARSZTFEJLŐDÉS XIX. Szombathely, 2014. pp. 137-146. A BÜKKI KARSZTVÍZSZINT ÉSZLELŐ RENDSZER KERETÉBEN GYŰJTÖTT HIDROMETEOROLÓGIAI ADATOK ELEMZÉSE ANALYSIS OF HYDROMETEOROLIGYCAL DATA OF BÜKK WATER LEVEL
Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and
 1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1321 0.2.2 Species name Myotis emarginatus 0.2.3 Alternative species scientific name 0.2.4 Common name csonkafülű denevér 1. National Level 1.1 Maps 1.1.1 Distribution
0.1 Member State HU 0.2.1 Species code 1321 0.2.2 Species name Myotis emarginatus 0.2.3 Alternative species scientific name 0.2.4 Common name csonkafülű denevér 1. National Level 1.1 Maps 1.1.1 Distribution
SAJTÓKÖZLEMÉNY Budapest 2011. július 13.
 SAJTÓKÖZLEMÉNY Budapest 2011. július 13. A MinDig TV a legdinamikusabban bıvülı televíziós szolgáltatás Magyarországon 2011 elsı öt hónapjában - A MinDig TV Extra a vezeték nélküli digitális televíziós
SAJTÓKÖZLEMÉNY Budapest 2011. július 13. A MinDig TV a legdinamikusabban bıvülı televíziós szolgáltatás Magyarországon 2011 elsı öt hónapjában - A MinDig TV Extra a vezeték nélküli digitális televíziós
Suppl. Materials. Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids. Germany
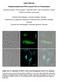 Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
7 th Iron Smelting Symposium 2010, Holland
 7 th Iron Smelting Symposium 2010, Holland Október 13-17 között került megrendezésre a Hollandiai Alphen aan den Rijn városában található Archeon Skanzenben a 7. Vasolvasztó Szimpózium. Az öt napos rendezvényen
7 th Iron Smelting Symposium 2010, Holland Október 13-17 között került megrendezésre a Hollandiai Alphen aan den Rijn városában található Archeon Skanzenben a 7. Vasolvasztó Szimpózium. Az öt napos rendezvényen
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1084 0.2.2 Species name Osmoderma eremita 0.2.3 Alternative species scientific name 0.2.4 Common name remetebogár 1. National Level 1.1 Maps 1.1.1 Distribution Map
0.1 Member State HU 0.2.1 Species code 1084 0.2.2 Species name Osmoderma eremita 0.2.3 Alternative species scientific name 0.2.4 Common name remetebogár 1. National Level 1.1 Maps 1.1.1 Distribution Map
Using the CW-Net in a user defined IP network
 Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 4029 0.2.2 Species name Chondrosoma fiduciarium 0.2.3 Alternative species scientific name 0.2.4 Common name magyar ősziaraszoló 1. National Level 1.1 Maps 1.1.1 Distribution
0.1 Member State HU 0.2.1 Species code 4029 0.2.2 Species name Chondrosoma fiduciarium 0.2.3 Alternative species scientific name 0.2.4 Common name magyar ősziaraszoló 1. National Level 1.1 Maps 1.1.1 Distribution
Computer Architecture
 Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
The beet R locus encodes a new cytochrome P450 required for red. betalain production.
 The beet R locus encodes a new cytochrome P450 required for red betalain production. Gregory J. Hatlestad, Rasika M. Sunnadeniya, Neda A. Akhavan, Antonio Gonzalez, Irwin L. Goldman, J. Mitchell McGrath,
The beet R locus encodes a new cytochrome P450 required for red betalain production. Gregory J. Hatlestad, Rasika M. Sunnadeniya, Neda A. Akhavan, Antonio Gonzalez, Irwin L. Goldman, J. Mitchell McGrath,
ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN
 ÉRETTSÉGI VIZSGA 2008. május 26. ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA 2008. május 26. 8:00 Az írásbeli vizsga időtartama: 180 perc Pótlapok száma Tisztázati Piszkozati OKTATÁSI
ÉRETTSÉGI VIZSGA 2008. május 26. ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA 2008. május 26. 8:00 Az írásbeli vizsga időtartama: 180 perc Pótlapok száma Tisztázati Piszkozati OKTATÁSI
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1214 0.2.2 Species name Rana arvalis 0.2.3 Alternative species Rana arvalis wolterstorffi scientific name 0.2.4 Common name hoszúlábú mocsári béka 1. National Level
0.1 Member State HU 0.2.1 Species code 1214 0.2.2 Species name Rana arvalis 0.2.3 Alternative species Rana arvalis wolterstorffi scientific name 0.2.4 Common name hoszúlábú mocsári béka 1. National Level
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1337 0.2.2 Species name Castor fiber 0.2.3 Alternative species scientific name 0.2.4 Common name európai hód 1. National Level 1.1 Maps 1.1.1 Distribution Map Yes
0.1 Member State HU 0.2.1 Species code 1337 0.2.2 Species name Castor fiber 0.2.3 Alternative species scientific name 0.2.4 Common name európai hód 1. National Level 1.1 Maps 1.1.1 Distribution Map Yes
Utasítások. Üzembe helyezés
 HASZNÁLATI ÚTMUTATÓ Üzembe helyezés Utasítások Windows XP / Vista / Windows 7 / Windows 8 rendszerben történő telepítéshez 1 Töltse le az AORUS makróalkalmazás telepítőjét az AORUS hivatalos webhelyéről.
HASZNÁLATI ÚTMUTATÓ Üzembe helyezés Utasítások Windows XP / Vista / Windows 7 / Windows 8 rendszerben történő telepítéshez 1 Töltse le az AORUS makróalkalmazás telepítőjét az AORUS hivatalos webhelyéről.
Contact us Toll free (800) fax (800)
 Table of Contents Thank you for purchasing our product, your business is greatly appreciated. If you have any questions, comments, or concerns with the product you received please contact the factory.
Table of Contents Thank you for purchasing our product, your business is greatly appreciated. If you have any questions, comments, or concerns with the product you received please contact the factory.
A magyarországi Gauss-Krüger-vetületû katonai topográfiai térképek dátumparaméterei
 A magyarországi Gauss-Krüger-vetületû katonai topográfiai térképek dátumparaméterei Timár Gábor 1 Kubány Csongor 2 Molnár Gábor 1 1ELTE Geofizikai Tanszék, Ûrkutató Csoport 2Honvédelmi Minisztérium, Térképészeti
A magyarországi Gauss-Krüger-vetületû katonai topográfiai térképek dátumparaméterei Timár Gábor 1 Kubány Csongor 2 Molnár Gábor 1 1ELTE Geofizikai Tanszék, Ûrkutató Csoport 2Honvédelmi Minisztérium, Térképészeti
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1353 0.2.2 Species name Canis aureus 0.2.3 Alternative species scientific name 0.2.4 Common name aranysakál 1. National Level 1.1 Maps 1.1.1 Distribution Map Yes
0.1 Member State HU 0.2.1 Species code 1353 0.2.2 Species name Canis aureus 0.2.3 Alternative species scientific name 0.2.4 Common name aranysakál 1. National Level 1.1 Maps 1.1.1 Distribution Map Yes
Választási modellek 3
 Választási modellek 3 Prileszky István Doktori Iskola 2018 http://www.sze.hu/~prile Forrás: A Self Instructing Course in Mode Choice Modeling: Multinomial and Nested Logit Models Prepared For U.S. Department
Választási modellek 3 Prileszky István Doktori Iskola 2018 http://www.sze.hu/~prile Forrás: A Self Instructing Course in Mode Choice Modeling: Multinomial and Nested Logit Models Prepared For U.S. Department
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel
 Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN
 ÉRETTSÉGI VIZSGA 2013. május 23. ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA 2013. május 23. 8:00 Az írásbeli vizsga időtartama: 180 perc Pótlapok száma Tisztázati Piszkozati EMBERI
ÉRETTSÉGI VIZSGA 2013. május 23. ELEKTRONIKAI ALAPISMERETEK ANGOL NYELVEN KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA 2013. május 23. 8:00 Az írásbeli vizsga időtartama: 180 perc Pótlapok száma Tisztázati Piszkozati EMBERI
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 1304 0.2.2 Species name Rhinolophus ferrumequinum 0.2.3 Alternative species scientific name 0.2.4 Common name nagy patkósdenevér 1. National Level 1.1 Maps 1.1.1
0.1 Member State HU 0.2.1 Species code 1304 0.2.2 Species name Rhinolophus ferrumequinum 0.2.3 Alternative species scientific name 0.2.4 Common name nagy patkósdenevér 1. National Level 1.1 Maps 1.1.1
DECLARATION OF PERFORMANCE No. GST REV 1.03 According to Construction Products Regulation EU No. 305/2011
 DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
ANNEX V / V, MELLÉKLET
 ANNEX V / V, MELLÉKLET VETERINARY CERTIFICATE EMBRYOS OF DOMESTIC ANIMALS OF THE BOVINE SPECIES FOR IMPORTS COLLECTED OR PRODUCED BEFORE 1 st JANUARY 2006 ÁLLAT-EGÉSZSÉGÜGYI BIZONYÍTVÁNY 2006. JANUÁR 1-JE
ANNEX V / V, MELLÉKLET VETERINARY CERTIFICATE EMBRYOS OF DOMESTIC ANIMALS OF THE BOVINE SPECIES FOR IMPORTS COLLECTED OR PRODUCED BEFORE 1 st JANUARY 2006 ÁLLAT-EGÉSZSÉGÜGYI BIZONYÍTVÁNY 2006. JANUÁR 1-JE
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 4020 0.2.2 Species name Pilemia tigrina 0.2.3 Alternative species scientific name 0.2.4 Common name atracél cincér 1. National Level 1.1 Maps 1.1.1 Distribution Map
0.1 Member State HU 0.2.1 Species code 4020 0.2.2 Species name Pilemia tigrina 0.2.3 Alternative species scientific name 0.2.4 Common name atracél cincér 1. National Level 1.1 Maps 1.1.1 Distribution Map
AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN
 Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella
 A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella Országos Meteorológiai Szolgálat Hungarian Meteorological Service KRITéR
A jövőbeli hatások vizsgálatához felhasznált klímamodell-adatok Climate model data used for future impact studies Szépszó Gabriella Országos Meteorológiai Szolgálat Hungarian Meteorological Service KRITéR
2016 év eleji ajánlatok. Speciális áraink 2016 május 31-ig érvényesek!
 2016 év eleji ajánlatok Speciális áraink 2016 május 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
2016 év eleji ajánlatok Speciális áraink 2016 május 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
First experiences with Gd fuel assemblies in. Tamás Parkó, Botond Beliczai AER Symposium 2009.09.21 25.
 First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
First experiences with Gd fuel assemblies in the Paks NPP Tams Parkó, Botond Beliczai AER Symposium 2009.09.21 25. Introduction From 2006 we increased the heat power of our units by 8% For reaching this
Téli kedvezményes ajánlataink
 Téli kedvezményes ajánlataink Ajánlataink 2017 január 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
Téli kedvezményes ajánlataink Ajánlataink 2017 január 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
FÖLDRAJZ ANGOL NYELVEN GEOGRAPHY
 Földrajz angol nyelven középszint 0513 ÉRETTSÉGI VIZSGA 2005. május 18. FÖLDRAJZ ANGOL NYELVEN GEOGRAPHY KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA STANDARD LEVEL WRITTEN EXAMINATION Duration of written examination:
Földrajz angol nyelven középszint 0513 ÉRETTSÉGI VIZSGA 2005. május 18. FÖLDRAJZ ANGOL NYELVEN GEOGRAPHY KÖZÉPSZINTŰ ÍRÁSBELI VIZSGA STANDARD LEVEL WRITTEN EXAMINATION Duration of written examination:
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 4110 0.2.2 Species name Pulsatilla pratensis ssp. hungarica 0.2.3 Alternative species Pulsatilla flavescens scientific name 0.2.4 Common name magyar kökörcsin 1.
0.1 Member State HU 0.2.1 Species code 4110 0.2.2 Species name Pulsatilla pratensis ssp. hungarica 0.2.3 Alternative species Pulsatilla flavescens scientific name 0.2.4 Common name magyar kökörcsin 1.
Szoftver-technológia II. Tervezési minták. Irodalom. Szoftver-technológia II.
 Tervezési minták Irodalom Steven R. Schach: Object Oriented & Classical Software Engineering, McGRAW-HILL, 6th edition, 2005, chapter 8. E. Gamma, R. Helm, R. Johnson, J. Vlissides:Design patterns: Elements
Tervezési minták Irodalom Steven R. Schach: Object Oriented & Classical Software Engineering, McGRAW-HILL, 6th edition, 2005, chapter 8. E. Gamma, R. Helm, R. Johnson, J. Vlissides:Design patterns: Elements
Report on the main results of the surveillance under article 11 for annex II, IV and V species (Annex B)
 0.1 Member State HU 0.2.1 Species code 4011 0.2.2 Species name Bolbelasmus unicornis 0.2.3 Alternative species scientific name 0.2.4 Common name szarvas álganéjtúró 1. National Level 1.1 Maps 1.1.1 Distribution
0.1 Member State HU 0.2.1 Species code 4011 0.2.2 Species name Bolbelasmus unicornis 0.2.3 Alternative species scientific name 0.2.4 Common name szarvas álganéjtúró 1. National Level 1.1 Maps 1.1.1 Distribution
