Salt-Enabled Visual Detection of DNA
|
|
|
- Eszter Kissné
- 6 évvel ezelőtt
- Látták:
Átírás
1 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Supporting Information for Salt-Enabled Visual Detection of DNA Xilong Wang, Xun Chen, Sha Sun, Feifei Zhang, and Jin Zhu* Department of Polymer Science and Engineering, School of Chemistry and Chemical Engineering, State Key Laboratory of Coordination Chemistry, Nanjing National Laboratory of Microstructures, Nanjing University, Nanjing , China *Corresponding author. Phone: ; Fax: ;
2 Table of Contents 1. Materials, facilities, and measurements...s3-s4 2. DNA and RNA sequence information...s5-s11 3. Preparation of self-assembled DNA nanostructure...s12-s16 4. Stability of Cuboid1...S17-S18 5. Preparation of DNA-modified glass slide... S19 6. Detection of DNA1 with Cuboid1... S20 7. Contact angle measurement... S21 8. Attempted detection of DNA1 with Probe1A or Probe1B mixture...s22-s23 9. Differentiation of DNA1 from DNA strands with single-base mismatches... S Two-target detection... S RNA detection... S DNA detection through LCR-rolling circle amplification (LCR- RCA) protocol...s27-s References... S30 S2
3 1. Materials, facilities, and measurements Hydrogen peroxide (30 wt%, AR grade) was purchased from Yonghua Chemical Technology (Jiangsu) Co. Ltd. Concentrated sulfuric acid (95-98%), hydrochloric acid (36-38 wt%), ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA, 99.0%), boric acid (99.5%), and ammonia solution (25-28 wt%) were purchased from Nanjing Chemical Reagent Co. Ltd. 3-(2-Aminoethylamino)propyltrimethoxysilane (EDAS, 96%), hexanoic anhydride (HA, 98%), sodium dihydrogen phosphate dehydrate (99%), sodium hydrogen phosphate (anhydrous, 99.0%), and sodium chloride (99%) were purchased from Alfa Aesar. Ethanol (99.7%) was from Shanghai Titanchem Co. Ltd. Succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC, 98% HPLC) was purchased from Meyer Chemical Technology Co. Ltd. Glacial acetic acid (99.8%) was from Acros Organics. 1-Hexanethiol (HT, 96%) was from Adamas-beta. Tris(hydroxymethyl)aminomethane (Tris, 99.8%) was purchased from Nanjing Biosky Sci. & Tech. Co. Ltd. NaNO 3 (96%) was from Xilong Chemical Co. Ltd. Magnesium chloride hexahydrate (99.0%) was purchased from Sigma. Sodium dodecyl sulfate (CP, SDS), dimethyl sulfoxide (DMSO, 99.0%), pyridine (99.5%), and N,N -dimethylformamide (DMF, 99.5%) were obtained from Sinopharm Chemical Reagent Co. Ltd. DMSO, pyridine, and DMF were distilled over CaH 2 prior to use, and other reagents were used as received. Taq DNA ligase was from New England Biolabs. φ29 DNA polymerase was from Thermo Scientific. E coli. Exonuclease I, E coli. exonuclease III, dntps (10mM), DL2000 DNA marker, and 20 bp DNA ladder marker were purchased from Takara Biotechnology (Dalian) Co. Ltd. Diethylpyrocarbonate (DEPC) and ethidium bromide were purchased from Sangon Biotech (Shanghai) Co. Ltd. Freeze N Squeeze column was purchased from Bio-Rad Laboratories, Inc. Nano-pure water (18.2 MΩ cm), purified by Sartorius Arium 611 system, was used throughout the experiment. All the DNA molecules (Tables S1 and S3-S5) were custom-synthesized by Sangon Biotech (Shanghai) Co. Ltd. RNA (Table S2) was custom-synthesized by Takara Biotechnology (Dalian) Co. Ltd. S3
4 Ligase chain reaction (LCR) and the preparation of self-assembled DNA nanostructure were performed on a GeneAmp PCR system 9700 from Applied Biosystems. Contact angle measurement was performed on a KSV CAM200 optical contact angle meter. Gel electrophoresis was performed on a Bio-Rad system using 2% agarose gel or 20% polyacrylamide. Photographs were taken using Canon DIGITAL IXUS 950IS digital camera. S4
5 2. DNA and RNA sequence information Tables S1. DNA symbol and sequence information Description of DNA Capture1 DNA1 Probe1 Probe1A DNA1-SBM1 DNA1-SBM2 DNA1-SBM3 DNA1-SBM4 Capture2 DNA2 Probe2 RCA1 PDNA1 AMPDNA1A AMPDNA1B Sequence a 5 -ATTTAACAATAATCCTTTTTTTTTT-SH-3 5 -GGATTATTGTTAAATATTGATAAGGAT-3 5 -TTTTTTTTTTATCCTTATCAAT-3 5 -ATCCTTATCAAT-3 5 -GGATTATCGTTAAATATTGATAAGGAT-3 5 -GGATTATTATTAAATATTGATAAGGAT-3 5 -GGATTATTCGTTAAATATTGATAAGGAT-3 5 -GGATTAT GTTAAATATTGATAAGGAT-3 5 -TATCATACTAACATATTTTTTTTTT-SH-3 5 -TATGTTAGTATGATATAGGAATAGTTA-3 5 -TTTTTTTTTTTAACTATTCCTA-3 5 -HS-AAAAAAAAAATCATCGAGGAAGTGGCAG TC-3 5 -(P)-TTAACAATAATGGTCTGCCCTGACTGCCAC TTCCTCGATGATCCTCGATGTCCTTATCAATAT-3 5 -ATTATTGTTAA-3 5 -(P)-ATATTGATAAGG-3 a (P) denotes the presence of a phosphate group. Tables S2. RNA symbol and sequence information Description of RNA RNA1 Sequence 5 -GGAUUAUUGUUAAAUAUUGAUAAGGAU-3 S5
6 Tables S3. DNA sequences for making Cuboid1 and Cuboid2 DNA strand Sequence (5 3 ) 1 CATAATACTTTTTTTTTTTTTTTTGGCTAGTAGATTGTTTGG AGTCAG 2 GCCCTTCCTTTTTTTTTTTTTTTTGGAGACCAAAGAATGTT GAGCAGG 3 CCCGCCCCTTTTTTTTTTTTTTTTGGGGACCGCGAGATATA AGACGTA 4 CTCCGGCCTTCTGATCCCCGCTGCCTGCCACCGAGCGCTG AGAACCGT 5 TACTACGTTCCCTCAACGGCTTACGCTCCGCAGTCGTGGC ATTACGAT 6 CAATTCTCAATATCCATAAATACCACATCCCGGCTGAGCC GAGACGTG 7 TGATTCCGCCACAAACGGCTGCAAGTAGCAAGATGCTGA GTGCCGTAT 8 TTCACAACCTAAGACCGTAGACGTTAGGCGAAAAGTCGA AAAAAATGA 9 TAAAACCTCGGGCCCCGGGCAGCTGGCGGCGAACGGGA GTCCGGCGAT 10 GCGGCCCACAATTTCCAGCATTCCTTTTTTTTTTTTTTTTG GAACTTA 11 ATTTAGCCCGGAGCCCTGTGCTCCTTTTTTTTTTTTTTTTG GTATTGT 12 TTAGAACTCCGATACCCGATATCCTTTTTTTTTTTTTTTTG GGTCCGG 13 CAGCCTTTCCTGTATAATGGCTAAGGGGCCAAAGGATCGT GAGGTCAT 14 TACGCCTATTCCAAATGTTCTGAGGGCCGGTGAAATGTGA GGCATAAA 15 CCTACCGACTCATAATTGGCAATAAGTTAAAAGCGGCGGG GTCTAGGA 16 TTTTTTTTCACCCAGTGCCCTATGCCCTTGATGGGGCGCT GCAGGATG 17 TTTTTTTTATACCATACAACTCATGTTTCCTACGCAGGGAG AGTAGAA 18 TTTTTTTTCCGCGGCTCCCAGCCCACTGACCCAGCTAAGC CAGGTCAG 19 CCATGCGTCCTACAGCCTGCGACCTACGACCTGTATTCAG GAACTCAG 20 CACCGCGCGCTTTATACACTTTACTTTTTTACAACGGGCTT GGGACCA 21 CCACTATTTTCGGTCACCTGAACCTTAAGTCCGCACGGGG AACACGAT S6
7 22 AAACAATCTACTAGCCGGAGTACAGGTGGCAGGCAGCGG GGATCAGAA 23 TATTGCCATACGTCTTATATCTCGCGGTCCCCGGGTTCCGC GGGATGT 24 TGTCACATCACGTCTCTGCCAGGGATTATGAGTCGGTAGG CGGTGATT 25 AGCGCCCCGTACTACATACATCCTACGGTTCTGACGGTAC TATACAGG 26 TTAATTCCCTTGCTCCGGGTGAATTGCGGGGCACCCAAG GACAGCGGA 27 ACCCCTGATTACCTGTCACCCGTAATCCGCCCGGTGTCAA CGACCCGG 28 GTCGGCACCAACATCCGGATAACGGTGACTGGTGGCTGG TCGTGATTT 29 CAATTGCCGCTAGCCCCCGAGTCCCGCACGCTGCGTGGA TTGGACGGG 30 CCCGGCCACCGGACCCGGGGGCCTGGGGCCCGAGGTTTT AGAGGTCCG 31 CTGAATACCTATGACCCGCAGCCCTAAGTTCCGCGCAAAT GTTTGTGG 32 CTCAGCATCTTGCTACTGGGCCGCAGGTCGTAGGTCGCA GGCTGTAGG 33 GCTTGCAAATCGCCGGACTCCCGTTCGCCGCCAGTTCTA AGGACTTAA 34 ACGATCCTTTGGCCCCGGCCGGAGATCAAGGGCATAGGG CACTGGGTG 35 CCTTAAACTCCTAGACCCCGCCGCTTTTAACTGAGAATTG GGGTCAGT 36 TATCACCCCTATCTCCTGGCCGAGGATTTTAGGGTCGCGG GTAGCGTG 37 CACGCCCAGCTATCCGTTACCCGGTCTTCTGGGGGCGGC AGCAATGTA 38 GCTATTGCATCGTGTTTGGAATGGGATCTGGGAACTAGGG ATCCGGTA 39 TAACCACAGTCTTTACACTAATCTCTGAGTTCCGGCTAAT GATGGCAG 40 TGTACTCCTTTTTTTT 41 TACTGCCCTTTTTTTT 42 CGGAACCCTTTTTTTT 43 GGGGCGGGAATCTAGG 44 CAGCGCTCCAAGTCCC 45 GGGCTGTGGAGCATTG 46 GCTTGTCCATCGGCCC 47 GGAATGCTGGAAATTG S7
8 48 TTTTTTTTGGTTTGTA 49 TTTTTTTTGGAAAACG 50 TTTTTTTTGGGATACT 51 AGCTGCCCAGTATCCC 52 ATTCACCCTTTTTTTTTTTTTTTTGGGACTTG 53 CACAGCCCTTTTTTTTTTTTTTTTGGGCGGAT 54 AGGCCCCCTTTTTTTTTTTTTTTTGGGCCGAT 55 ACATTCTTTGGTCTCCGGGCAGTATGCGGAGC 56 TTGCAGCCTACAAACCGGAGCACAGGGCTCCG 57 ACGTCTACCGTTTTCCGGATATCGGGTATCGG 58 CCCTGGCACCTAGATTTACGGGTGACAGGTAA 59 CCTTGGGTGCCCCGCAGGACAAGCAGCGTGCG 60 CGTTATCCCAATGCTCGGGCTGCGGGTCATAG 61 TTAGCCATCTGACTCCGTAAGCCGTTGAGGGA 62 CTCAGAACCCTGCTCAGGTATTTATGGATATT 63 TTCGACTTTTCGCCTAGGCTAAATGTAAAAAA 64 CTCGGCCATCCGCTGTAGGATGTATGTAGTAC 65 ACCAGCCACCAGTCACTCAGGGGTCCAGAAGA 66 CCATTCCACGGACCTCGGACTCGGGGGCTAGC 67 TCACATTTCACCGGCCACGTAGTATAGGAAAC 68 CATTCGTCATACGGCAGTAAAGTGTATAAAGC 69 CTTGACTTTCATTTTTGGTTCAGGTGACCGAA 70 TTAACATCAATCACCGCCGGGTAACGGATAGC 71 CCGCGACCCTAAAATCGGCAATTGGATACGTA 72 CATCTCACAAATCACGAGATTAGTGTAAAGAC 73 TATGTCTCATGACCTCATGAGTTGTATGGTAT 74 CCAAGTTCTTTATGCCGGGCTGGGAGCCGCGG 75 GTCTTGGCCTGACCTGGATGTTAAGTGACGGA 76 GTACCTCGCATCCTGCATGGTTGAGTGAACGA 77 TGTGTCCACATTCAATGTGAGATGCAGGTTGC 78 ATCCGCGCTACGTATCGGCAGGAGCCAACGGT 79 TTCGCGTGCCACACGTACGCATGGCGTCGGGT 80 CAAACGCCTTGTGTCTGCGCGGTGTGTATTGA 81 CTTCCGATCGCCCGTAAATAGTGGTACGAAGT 82 CCGCTTCCTTTTTTTTTTTTTTTTGTGCTTGC 83 TTGACACCTTTTTTTTTTTTTTTTGGTTCGGA 84 ACGCGGCCTTTTTTTTTTTTTTTTGGTGAGGA 85 CCGCCTCCTTTTTTTTTTTTTTTTGGAGCAAG 86 TCGATCTCTTTTTTTTTTTTTTTTGGGACAAA 87 ATTTGCGCTTTTTTTTTTTTTTTTGAGGACAT 88 GCCACGACGCAAGCACGTATTATGGCGAAGAT 89 GGCTCAGCTCCGAACCGGAAGGGCTATGATAA 90 CATATCCCTCCTCACCGGAAGCGGGAGGATGG 91 CATCCGTCTTTGTCCCGGAGGCGGGTGCGAGA S8
9 92 ATTATCCTATGTCCTCGGCCGCGTGAGTTAGA 93 TAACTATTACAATACCGAGATCGAGGTCTTAG 94 CTCATTTCTTATCATAGGGATATGGAAACTAG 95 GTACCGTCATCTTCGCGGAATTAAACCGGGGT 96 AATTAGATCCGGGTCGAGGATAATGGATGTTG 97 TACTACCTCCATCCTCGACGGATGAGTATATT 98 CCACAGATTCTAACTCAATAGTTAATGTCAGA 99 ATCCACGCTCTCGCACTGGCCGGGACGAAGGC 100 GATCCCGTATCGTAATGTCGTGGCATTACGAT 101 AACCCTTTACCCCGGTAGGTAGTAGTTGAAAG 102 TTAAATATCTAGTTTCATCTAATTGGTAGGGT 103 ATTTCGTGAATATACTATCTGTGGGGCAGATG 104 CCCCGTGCGCCTTCGTGTTGTGAAGTTCGAGC 105 AGCCCGTTTCTGACATCGGAATCAACAAGTCA 106 TTATTCTTCTTTCAACACGGGATCGGGCACAG 107 TGCCGCCCACCCTACCATGTGACAAGGTTTGA 108 CGCATCTACATCTGCCATATTTAAGTTTTCAG 109 CGTCCGTACCCGTCCAAAAGGGTTGGAGATAG 110 GGACTCTAGCTCGAACCACGAAATAGAAAGCA 111 ATTAGCCGTGACTTGTGTGCCGACAATGCAGT 112 TCCCTGCGCTGTGCCCAAAGGCTGGATAGAGA 113 GCTTAGCTTCAAACCTTAGGCGTAGTGTGGTG 114 CCTCTACCCTGAAAACAAGAATAAGGTATGGG 115 CCAGCACCTGCTTTCTTACGGACGCTGACGTA 116 TTACAGCTACTGCATTTAGATGCGGGCGCGAG 117 TCGTGCCTTGGTCCCATAGAGTCCAGGGCAAG 118 CGCTACAACACCACACGGTAGAGGAAGAGAGG 119 TCAACCATTCTCTATCGGGTGATAGTGATTAA 120 ATATAACGTACATTGCAGCTGTAAATTGAATG 121 CGATTACTCCCATACCGGTGCTGGTGACGAGG 122 CGATCATGCTCGCGCCAGGCACGAGAACGTGG 123 CTCCTGCCTACGTCAGGCAATAGCAAACGAAT 124 AACTCCACTTCTACTCTTGTAGCGAGGCAACG 125 CATTTATCTTAATCACAGTAATCGCTGCGAAA 126 TACTTCCACCTCTCTTCGTTATATGCGATGTC 127 GCACTCGCCCTCGTCACATGATCGCGGAGACA 128 CAATATGTATTCGTTTAACATGGGCTCGGGTT 129 GTTCAGACCCACGTTCTTGCGGATACAAGAGC S9
10 Tables S4. Probe DNA sequences for making Cuboid1 DNA strand Sequence (5 3 ) Probe1B-1 TTTTTTTTTTTCGCAGGTGGAGTTTTTTTTTTTTATCCTTA TCAAT Probe1B-2 TTTTTTTTTGTCTCCGTGGAAGTATTTTTTTTTTATCCTTA TCAAT Probe1B-3 TTTTTTTTAACCCGAGGCGAGTGCTTTTTTTTTTATCCTT ATCAAT Probe1B-4 TTTTTTTTGACATCGCGCCAAGACTTTTTTTTTTATCCTTA TCAAT Probe1B-5 TTTTTTTTACCGTTGGGATAAATGTTTTTTTTTTATCCTTA TCAAT Probe1B-6 TTTTTTTTGCTCTTGTTGGACACATTTTTTTTTTATCCTTA TCAAT Probe1B-7 ATCCGCAACTGCCATCGACGAATGACGTGTGGCACGCG AATTTTTTTTTTATCCTTATCAAT Probe1B-8 CCCATGTTCTTGCCCTAAGTCAAGAGACACAAGGCGTTT GTTTTTTTTTTATCCTTATCAAT Probe1B-9 CCCTAGTTCCCAGATCTTGCAAGCTACGGGCGATCGGAA GTTTTTTTTTTATCCTTATCAAT Probe1B-10 TTTTTTTTTCGTTCACGAGACATATTTTTTTTTTATCCTTA TCAAT Probe1B-11 TTTTTTTTCGTTGCCTGAACTTGGTTTTTTTTTTATCCTTA TCAAT Probe1B-12 TTTTTTTTTCCGTCACGTTTAAGGTTTTTTTTTTATCCTTA TCAAT Probe1B-13 TTTTTTTTCACGCTACCGAGGTACTTTTTTTTTTATCCTTA TCAAT Probe1B-14 TTTTTTTTGCAACCTGTGGGCGTGTTTTTTTTTTATCCTTA TCAAT Probe1B-15 TTTTTTTTTACCGGATGCGCGGATTTTTTTTTTTATCCTTA TCAAT Probe1B-16 TTTTTTTTACTTCGTAACATATTGTTTTTTTTTTATCCTTAT CAAT Probe1B-17 TTTTTTTTTCAATACAGTCTGAACTTTTTTTTTTATCCTTA TCAAT Probe1B-18 TTTTTTTTACCCGACGTGTGGTTATTTTTTTTTTATCCTTA TCAAT S10
11 Tables S5. Probe DNA sequences for making Cuboid2 DNA strand Sequence (5 3 ) Probe2-1 TTTTTTTTTTTCGCAGGTGGAGTTTTTTTTTTTTTAACTATTC CTA Probe2-2 TTTTTTTTTGTCTCCGTGGAAGTATTTTTTTTTTTAACTATTC CTA Probe2-3 TTTTTTTTAACCCGAGGCGAGTGCTTTTTTTTTTTAACTATT CCTA Probe2-4 TTTTTTTTGACATCGCGCCAAGACTTTTTTTTTTTAACTATT CCTA Probe2-5 TTTTTTTTACCGTTGGGATAAATGTTTTTTTTTTTAACTATTC CTA Probe2-6 TTTTTTTTGCTCTTGTTGGACACATTTTTTTTTTTAACTATTC CTA Probe2-7 ATCCGCAACTGCCATCGACGAATGACGTGTGGCACGCGAA TTTTTTTTTTTAACTATTCCTA Probe2-8 CCCATGTTCTTGCCCTAAGTCAAGAGACACAAGGCGTTTG TTTTTTTTTTTAACTATTCCTA Probe2-9 CCCTAGTTCCCAGATCTTGCAAGCTACGGGCGATCGGAAG TTTTTTTTTTTAACTATTCCTA Probe2-10 TTTTTTTTTCGTTCACGAGACATATTTTTTTTTTTAACTATTC CTA Probe2-11 TTTTTTTTCGTTGCCTGAACTTGGTTTTTTTTTTTAACTATTC CTA Probe2-12 TTTTTTTTTCCGTCACGTTTAAGGTTTTTTTTTTTAACTATTC CTA Probe2-13 TTTTTTTTCACGCTACCGAGGTACTTTTTTTTTTTAACTATT CCTA Probe2-14 TTTTTTTTGCAACCTGTGGGCGTGTTTTTTTTTTTAACTATT CCTA Probe2-15 TTTTTTTTTACCGGATGCGCGGATTTTTTTTTTTTAACTATTC CTA Probe2-16 TTTTTTTTACTTCGTAACATATTGTTTTTTTTTTTAACTATTC CTA Probe2-17 TTTTTTTTTCAATACAGTCTGAACTTTTTTTTTTTAACTATTC CTA Probe2-18 TTTTTTTTACCCGACGTGTGGTTATTTTTTTTTTTAACTATTC CTA S11
12 3. Preparation of self-assembled DNA nanostructure Design of DNA nanostructure. The 6H (helices)/6h/64bp (base pairs) nanostructure (Cuboid1 and Cuboid2) was designed according to published methods developed previously, S1 assisted by program Sequin S2 (Cuboid1: DNA strands from Tables S3 and S4; Cuboid2: DNA strands from Tables S3 and S5). The 6H/6H/64BP nanostructure is a 6H by 6H by 64BP cuboid and 15 nm 15 nm 21.5 nm in size, with the addition of 8nt (nucleotides) repeat T at the head and tail of each helix to enhance its stability except for the Probe1- or Probe2-linking 18 helices (with a 10nt repeat T linker at the tail). Preparation of DNA nanostructure. 147 DNA strands were mixed and freeze-dried. The dried DNA strand mixture was dissolved in 0.5 TE buffer (5 mm Tris, 1 mm EDTA, ph 8.0) supplemented with 40 mm MgCl 2 to a final concentration of 200 nm per strand. The strand mixture was then annealed in a PCR thermocycler by a fast linear cooling step from 90 o C to 61 o C over 2.5 h, and subsequently from 60 o C to 24 o C over 74 h. Purification of DNA nanostructure. Annealed samples were loaded onto 2% agarose gel in 0.5 TBE buffer (44.6 mm Tris, 44.6 mm boric acid, 1 mm EDTA, ph 8.0) supplemented with 11 mm MgCl 2 and 0.5 μg/ml ethidium bromide. The gel was subjected to electrophoresis at 80 volts for 100 min in an ice water bath. Then, the target gel bands were visualized with ultraviolet light and excised. The excised bands were cut into small pieces and transferred into a Freeze N Squeeze column and the column was frozen at -20 o C for 5 min, then centrifuged at 7000 g for 5 min. Samples that were extracted through the column were collected and freeze-dried. The freeze-dried nanostructure was dissolved with water and the concentration of nanostructure was determined according to A 260 prior to use. Transmission electron microscopy (TEM) imaging. Unpurified or purified samples (2.5 µl) were loaded on a glass slide, and a carbon-coated TEM grid was placed on top of the samples for 2 min. The grid was then stained with 2% aqueous uranyl formate solution containing 25 mm NaOH for 2 min. The stained grid was washed S12
13 with water twice. Imaging was performed using a JEOL JEM-1011 operated at 100 kv. Fig. S1 3D model of the nanostructure 6H/6H/64BP. Fig. S2 Locations of the 18 helices containing Probe1 or Probe2. The 18 helices have been marked with white circles. S13
14 Fig. S3 Agarose gel electrophoresis diagram of the annealed Cuboid1 and Cuboid2. Lane M: DL2000 DNA marker; lane 1: annealed Cuboid1; lane 2: annealed Cuboid2. The bands marked in red represent self-assembled DNA nanostructures. Fig. S4 TEM image of the annealed Cuboid1. S14
15 Fig. S5 TEM image of the purified Cuboid1 as extracted from the band marked in the lane 1 of Fig. S3. Fig. S6 TEM image of the annealed Cuboid2. S15
16 Fig. S7 TEM image of the purified Cuboid2 as extracted from the band marked in the lane 2 of Fig. S3. S16
17 4. Stability of Cuboid1 Annealed Cuboid1 (6 μl) was separately challenged with 54 μl of buffer solution A, B, and C. Buffer solution A: 0.5 TE buffer supplemented with 40 mm MgCl 2. Buffer solution B: equal portions of 0.3 M PBS solution (0.3 M NaCl, 10 mm phosphate buffer, ph 7.0) and 5 TBE buffer supplemented with 110 mm MgCl 2. Buffer solution C: 5 TBE buffer supplemented with 55 mm MgCl 2. The diluted Cuboid1 (60 μl, in buffer A, B, and C) was divided into two equal portions, each of which (30 μl) was placed at 4 o C and 37 o C for 12 h, and then 6 μl of 6 loading buffer was placed in each solution. The mixture was loaded onto 2% native agarose gel in 0.5 TBE buffer supplemented with 11 mm MgCl 2 and 5 μg/ml ethidium bromide. The gel was subjected to electrophoresis at 80 volts for 60 min in an ice water bath. Annealed Cuboid1 (3 μl) was separately challenged with 27 μl of buffer solution B, C, and D. Buffer solution D: equal portions of 0.5 TE buffer supplemented with 40 mm MgCl 2 and 5 TBE buffer supplemented with 110 mm MgCl 2. The diluted Cuboid1 (30 μl, in buffer B, C, and D) was placed at 25 o C for 12 h, and then 6 μl of 6 loading buffer was placed in each solution. The mixture was loaded onto 2% native agarose gel in 0.5 TBE buffer (ph 8.0) supplemented with 11 mm MgCl 2 and 5 μg/ml ethidium bromide. The gel was subjected to electrophoresis at 80 volts for 60 min in an ice water bath. S17
18 Fig. S8 Stability of Cuboid1 in buffer solution A, B, and C at 4 o C and 37 o C. Agarose gel electrophoresis diagram showing stability of Cuboid1 in buffer solution A, B, and C at 4 o C and 37 o C after 12 h. Lane M: DL2000 DNA marker; lane 0: annealed Cuboid1; lane 1: annealed Cuboid1 in buffer solution A at 4 o C; lane 2: annealed Cuboid1 in buffer solution A at 37 o C; lane 3: annealed Cuboid1 in buffer solution B at 4 o C; lane 4: annealed Cuboid1 in buffer solution B at 37 o C; lane 5: annealed Cuboid1 in buffer solution C at 4 o C; lane 6: annealed Cuboid1 in buffer solution C at 37 o C. The bands marked in red represent Cuboid1. Fig. S9 Stability of Cuboid1 in buffer solution B, C, and D at 25 o C. Agarose gel electrophoresis diagram showing stability of Cuboid1 in buffer solution B, C and D at 25 o C after 12 h. Lane 1: annealed Cuboid1; lane 2: annealed Cuboid1 in buffer solution B at 25 o C; lane 3: annealed Cuboid1 in buffer solution C at 25 o C; lane 4: annealed Cuboid1 in buffer solution D at 25 o C. The bands marked in red represent Cuboid1. S18
19 5. Preparation of DNA-modified glass slide A glass slide was kept in a Piranha solution (hydrogen peroxide:concentrated sulfuric acid = 3:7, v/v) at 90 o C for 2 h. The glass slide was washed with copious amount of water and then placed in a mixture of hydrogen peroxide:ammonia solution:water (1:1:5, v/v/v) at 75 o C for 30 min. The glass slide was again washed with water and then placed in a mixture of hydrogen peroxide:hydrochloric acid:water (1:1:5, v/v/v) at 75 o C for 30 min. After this the glass slide was washed and incubated in an aqueous solution of EDAS (1% EDAS, v/v, 1 mm acetic acid) at room temperature for 30 min. After being washed with water, the EDAS-modified glass slide was N 2 -dried and then baked at 120 o C for 30 min. After being cooled to room temperature, the glass slide was immersed overnight in 5 mm DMSO solution of SMCC at a temperature of approximately or slightly above 25 o C. The glass slide was washed with copious amount of ethanol and water, and then dried by N 2, and then immersed in a mixture of DMF:pyridine:hexanoic anhydride (4:1:1, v/v/v) at 4 o C for 2 h. The SMCC-modified glass slide was washed with ethanol and water, and then N 2 -dried. Then 0.1 M PBS solution (0.1 M NaCl, 10 mm phosphate buffer, ph 7.0) of Capture1 (10 μm, 3 μl for each spot) was spotted onto surface of the SMCC-modified glass slide and the reaction was allowed to proceed at 37 o C for 8 h in a humidity chamber. After being washed with water and dried with N 2, the glass slide was immersed in an ethanol solution of 1 mm 1-hexanethiol at room temperature for 12 h. Finally the glass slide was washed with ethanol and water, and then N 2 -dried. The modification of Capture2 and RCA1 was performed in an analogous manner. S19
20 6. Detection of DNA1 with Cuboid1 A 0.3 M PBS solution (ph 7.0, containing 0.01 wt% SDS, 1.5 μl) of DNA1 was spotted onto a Capture1-modified glass slide. Then a 5 TBE buffer solution (1.5 μl) supplemented with 110 mm MgCl 2 of Cuboid1 (100 nm) was added and the solution was thoroughly mixed. The hybridization process was performed at 25 o C for 12 h in a humidity chamber and then each spot on the glass slide was washed once with 50 μl of 1 M PBN (1 M NaNO 3, 10 mm phosphate buffer, ph 7.0) supplemented with 11 mm MgCl 2 and 0.05 wt% SDS, followed by another three times wash with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and then the glass slide was placed in an oven at 40 o C for 4 min for the removal of water on the spots. The image was recorded using a Canon DIGITAL IXUS 950IS digital camera. For image acquisition, the glass slide was intentionally suspended at an angle of 45 degrees in the air with the black photography backdrop placed away from the glass slide. The visible light source was positioned at the lateral upper side of the glass slide surface and the digital camera was placed in in parallel with the glass slide. The image acquisition was performed in a high-quality ( pixels), lossy compression jpeg format. The utility of this format has enabled consistent, reproducible, and high-quality recording of macroscopic image under our experimental condition. The area of each salt spot on the glass slide in the image was calculated with the ImageJ software. The image was cut to the same size and the image type was changed from RGB color to 8-bit before the calculation. S20
21 7. Contact angle measurement A 0.3 M PBS solution (ph 7.0, containing 0.01 wt% SDS, 4.5 μl) containing no DNA or DNA1 (100 nm) was respectively spotted onto a Capture1-modified glass slide. Then a 5 TBE buffer solution (4.5 μl) supplemented with 110 mm MgCl 2 of Cuboid1 (100 nm) was added and the solution was thoroughly mixed. The hybridization process was performed at 25 o C for 12 h in a humidity chamber and then each spot on the glass slide was washed with 50 μl of 1 M PBN supplemented with 11 mm MgCl 2 and 0.05 wt% SDS, followed by another two times wash with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and finally each spot was washed with 50 μl of water, and then N 2 -dried. A 1 M PBN solution (3 μl) supplemented with 11 mm MgCl 2 was dropped onto the glass slide and contact angle was measured. Fig. S10 Contact angle measured at different locations of the glass slide. Macroscopic image of salt solution droplet showing the contact angle at different locations of the glass slide. From left to right: Capture1-free area, Capture1-modified area, Capture1-modified area after hybridization in the absence of DNA1, Capture1-modified area after hybridization in the presence of DNA1. Table S6. Contact angle at different locations of the glass slide. (ref. Fig. S10) Contact angle ( o ) Capture1- free area Capture1- modified area Capture1- modified area after hybridization in the absence of DNA1 Capture1- modified area after hybridization in the presence of DNA S21
22 8. Attempted detection of DNA1 with Probe1A or Probe1B mixture A 0.3 M PBS solution (ph 7.0, containing 0.01 wt% SDS, 1.5 μl) of DNA1 was spotted onto a Capture1-modified glass slide. Then a 5 TBE buffer solution (1.5 μl) supplemented with 110 mm MgCl 2 of Probe1A (1.8 μm) or Probe1B mixture (100 nm per Probe-1B strand; for sequence information, see Table S4) was added and the solution was thoroughly mixed. The hybridization process was performed at 25 o C for 12 h in a humidity chamber and then each spot on the glass slide was washed once with 50 μl of 1 M PBN supplemented with 11 mm MgCl 2 and 0.05 wt% SDS, followed by another three or four times wash with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and then the glass slide was placed in an oven at 40 o C for 4 min for the removal of water on the spots. S22
23 Fig. S11 Attempted DNA detection with Probe1A. A) Macroscopic image of a spot array on a glass slide for the attempted detection of DNA1 with Probe1A. DNA1 concentration (from left to right): 50 nm, 5 nm, and 0 M. The concentration of Probe1A is 900 nm. The glass slide was washed three times with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2. B) Macroscopic image of a spot array on a glass slide for the detection of DNA1 with Probe1A. DNA1 concentration (from left to right, from top to bottom): 500 nm, 100 nm, 50 nm, 10 nm, 5 nm, and 0 M. The concentration of Probe1A is 900 nm. The glass slide was four times washed with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2. Fig. S12 Attempted DNA detection with Probe1B mixture. A) Macroscopic image of a spot array on a glass slide for the detection of DNA1 with Probe1B mixture. DNA1 concentration (from left to right): 5 nm, 0 M, and 0 M. The concentration of Probe1B mixture is 50 nm (50 nm per Probe1B strand). The glass slide was washed three times with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2. B) Macroscopic image of a spot array on a glass slide for the detection of DNA1 with Probe1B mixture. DNA1 concentration (from left to right, from top to bottom): 100 nm, 50 nm, 25 nm, 10 nm, 5 nm, and 0 M. The concentration of Probe1B mixture is 50 nm (50 nm per Probe1B strand). The glass slide was washed four times with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2. S23
24 9. Differentiation of DNA1 from DNA strands with single-base mismatches A 75 mm PBS solution (75 mm NaCl, 2.5 mm phosphate buffer, containing wt% SDS, ph 7.0, 3 μl) of either DNA1 or a DNA strand with single-base mismatch (DNA1-SBM1, DNA1-SBM2, DNA1-SBM3, DNA1-SBM4) (50 nm) was spotted onto a Capture1-modified glass slide. The hybridization process was performed at 25 o C for 1 h in a humidity chamber and then each spot on the glass slide was washed with 50 μl of 60 mm PBS solution (60 mm NaCl, 2 mm phosphate buffer, ph 7.0) three times and then dried by N 2. A 0.5 TBE buffer (1.5 μl) supplemented with 11 mm MgCl 2 of Cuboid1 (20 nm) was spotted onto the dried spot and the hybridization process was performed at 25 o C for 1 h in the humidity chamber. Each spot was washed with 50 μl of 7.5 mm PBN (7.5 mm NaNO 3, 0.75 mm phosphate buffer, ph 7.0) supplemented with 1.25 mm MgCl 2 and 0.05 wt% SDS, followed by another three times wash with 50 μl of pure 7.5 mm PBN (ph 7.0) supplemented with 1.25 mm MgCl 2. Finally, each spot was washed with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and then the glass slide was placed in an oven at 40 o C for 4 min for the removal of water on the spots. S24
25 10. Two-target detection In a typical two-target detection experiment, a glass slide was spotted with 4 spots each of Capture1 (top row) and Capture2 (bottom row). The four columns of spots were each tested for a sample containing no DNA target, containing DNA1 (100 nm), containing DNA2 (100 nm), and containing both DNA1 (100 nm) and DNA2 (100 nm), respectively. The solutions of Cuboid1 and Cuboid2 were added to the 4 spots on the top and bottom rows, respectively. The hybridization was performed in an analogous manner (volume and concentration of each component except for DNA in the hybridization solution, duration of hybridization, concentration of each component in the wash buffer, and wash step) as that for the detection of single target. S25
26 11. RNA detection RNA1 powder was dissolved with DEPC-treated 0.3 M PBS (0.01 wt% SDS, ph 7.0) and diluted to different concentrations. The freeze-dried Cuboid1 was dissolved with DEPC-treated water and diluted with DEPC-treated 5 TBE buffer solution (ph 8.0) supplemented with 110 mm MgCl 2 to 100 nm. The Capture1-modified glass slide was washed with DEPC-treated water and dried by N 2. A DEPC-treated 0.3M PBS solution (ph 7.0, containing 0.01 wt% SDS, 1.5 μl) of RNA1 was spotted onto the Capture1-modified glass slide. And then a DEPC-treated 5 TBE buffer solution (ph 8.0, 1.5 μl) supplemented with 110 mm MgCl 2 of Cuboid1 (100 nm) was added and the solution was thoroughly mixed. The hybridization process was performed at 25 o C for 4 h in a humidity chamber and then each spot on the glass slide was washed with 50 μl of 1 M PBN supplemented with 11 mm MgCl 2 and 0.05 wt% SDS, followed by another three times wash with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and then the glass slide was placed in an oven at 40 o C for 4 min for the removal of water on the spots. S26
27 12. DNA detection through LCR-rolling circle amplification (LCR- RCA) protocol In a typical DNA detection experiment, a 20 μl LCR solution containing PDNA1 (1 μm), AMPDNA1A (1 μm), AMPDNA1B (1 μm), Taq DNA ligase (1 U/μL), Taq DNA ligase reaction buffer, and DNA1 (various concentrations) was subjected to a thermal cycling treatment: 65 o C, 5 min/20 cycles of (65 o C, 30 sec/25 o C, 2 min/45 o C, 3 min)/65 o C, 7 min. Then the LCR mixture (20 μl) containing CPDNA1 was subjected to enzymatic digestion in a final volume of 23.2 μl solution (exonuclease I 0.2 U/μL, exonuclease III 1.6 U/μL, 0.5 μl each of the two 10 nuclease reaction buffer) at 37 o C for 1 h for the removal of single-strand DNA, DNA1, PDNA1, AMPDNA1A, AMPDNA1B, and ligation product from AMPDNA1A and AMPDNA1B. The exonucleases were heat inactivated by incubation at 80 o C for 15 min. A RCA reaction solution was prepared through the mixing of following solutions: 10 mm dntps (2 μl), 10 φ29 DNA polymerase reaction buffer (2 μl), and φ29 DNA polymerase (1 μl). The RCA reaction solution (0.65 μl) was added to LCR mixture (3 μl) containing CPDNA1 and the solution was thoroughly mixed. The above mixture (3 μl) was spotted onto the RCA1-modified glass slide. The RCA reaction process was allowed to perform at 37 o C for 5 h in a humidity chamber and then each spot on the glass slide was washed with 50 μl of 1 M PBN supplemented with 11 mm MgCl 2 and 0.05 wt% SDS, followed by another six times wash with 50 μl of pure 1 M PBN supplemented with 11 mm MgCl 2, and then the glass slide was placed in an oven at 40 o C for 4 min for the removal of water on the spots. S27
28 Fig. S13 Verification of the ligation reaction by Taq DNA ligase. Gel electrophoresis diagram showing the ligation of PDNA1 by Taq DNA ligase in the presence of DNA1. Lane M: DNA ladder marker; lanes 1 to 6: DNA1 concentration of 100 nm, 10 nm, 1 nm, 100 pm, 10 pm, 0 M; lane 7: PDNA1; lane 8: DNA1. The bands marked in red represent the ligation product CPDNA1. PDNA1 (1 μm), AMPDNA1A (1 μm), and AMPDNA1B (1 μm) were ligated by Taq DNA ligase (1 U/μL) in the presence of DNA1 (various concentrations) under LCR condition: 65 o C, 5 min/20 cycles of (65 o C, 30 sec/25 o C, 2 min/45 o C, 3 min)/65 o C, 7 min. S28
29 Fig. S14 Verification of enzyme digestion of exonuclease I and exonuclease III. Gel electrophoresis diagram showing the ligation of PDNA1 and enzymatic digestion with exonuclease I and exonuclease III. Lane M: DNA ladder marker; lane 1: ligation mixture (concentration of DNA1 is 100 nm) before enzymatic digestion; lane 2: ligation mixture (concentration of DNA1 is 100 nm) after enzymatic digestion; lane 3: ligation mixture (concentration of DNA1 is 10 nm) before enzymatic digestion; lane 4: ligation mixture (concentration of DNA1 is 10 nm) after enzymatic digestion. The bands marked in red represent the ligation product CPDNA1. S29
30 13. References S1. Y. Ke, L. L. Ong, W. M. Shih, P. Yin, Science, 2012, 338, S2. N. C. Seeman, J. Biomol. Struct. Dyn., 1990, 8, S30
Supporting Information
 Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Supporting Information Cell-free GFP simulations Cell-free simulations of degfp production were consistent with experimental measurements (Fig. S1). Dual emmission GFP was produced under a P70a promoter
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing MFMER slide-1
 Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Trinucleotide Repeat Diseases: CRISPR Cas9 PacBio no PCR Sequencing 2015 MFMER slide-1 Fuch s Eye Disease TCF 4 gene Fuchs occurs in about 4% of the US population. Leads to deteriorating vision without
Supporting Information
 Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supporting Information Paired design of dcas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains Yihao Zhang 1,2,8, Long Qian 4,8, Weijia Wei 1,3,7,8, Yu
Supplementary Table 1. Cystometric parameters in sham-operated wild type and Trpv4 -/- rats during saline infusion and
 WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
WT sham Trpv4 -/- sham Saline 10µM GSK1016709A P value Saline 10µM GSK1016709A P value Number 10 10 8 8 Intercontractile interval (sec) 143 (102 155) 98.4 (71.4 148) 0.01 96 (92 121) 109 (95 123) 0.3 Voided
A cell-based screening system for RNA Polymerase I inhibitors
 Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2019 Supporting Information A cell-based screening system for RNA Polymerase I inhibitors Xiao Tan,
University of Bristol - Explore Bristol Research
 Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Al-Salihi, S. A. A., Scott, T. A., Bailey, A. M., & Foster, G. D. (2017). Improved vectors for Agrobacterium mediated genetic manipulation of Hypholoma spp. and other homobasidiomycetes. Journal of Microbiological
Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and
 1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
1 2 3 4 5 Journal name: Applied Microbiology and Biotechnology Manuscript Title: Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic
Modular Optimization of Hemicellulose-utilizing Pathway in. Corynebacterium glutamicum for Consolidated Bioprocessing of
 [Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
[Supplementary materials] Modular Optimization of Hemicellulose-utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass Sung Sun Yim 1, Jae Woong Choi 1,
Contact us Toll free (800) fax (800)
 Table of Contents Thank you for purchasing our product, your business is greatly appreciated. If you have any questions, comments, or concerns with the product you received please contact the factory.
Table of Contents Thank you for purchasing our product, your business is greatly appreciated. If you have any questions, comments, or concerns with the product you received please contact the factory.
Whole mount in situ hybridization, sectioning, and immunostaining Ribonuclease protection assay (RPA) Collagen gel tube formation assay
 Whole mount in situ hybridization, sectioning, and immunostaining In situ hybridization for tie-1 and lncrna tie-1as and sectioning of ISH stained embryos were performed according to previously published
Whole mount in situ hybridization, sectioning, and immunostaining In situ hybridization for tie-1 and lncrna tie-1as and sectioning of ISH stained embryos were performed according to previously published
Construction of a cube given with its centre and a sideline
 Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Transformation of a plane of projection Construction of a cube given with its centre and a sideline Exercise. Given the center O and a sideline e of a cube, where e is a vertical line. Construct the projections
Új temékek az UD- GenoMed Kft. kínálatában!
 Új temékek az UD- GenoMed Kft. kínálatában! Szolgáltatásaink: Medical Genomic Technologies Kft. Betegtoborzás és biobanking Bioinformatika o Adatelemzés/adatbányászás o Integrált adatbázis készítés Sejtvonal
Új temékek az UD- GenoMed Kft. kínálatában! Szolgáltatásaink: Medical Genomic Technologies Kft. Betegtoborzás és biobanking Bioinformatika o Adatelemzés/adatbányászás o Integrált adatbázis készítés Sejtvonal
Forensic SNP Genotyping using Nanopore MinION Sequencing
 Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
Forensic SNP Genotyping using Nanopore MinION Sequencing AUTHORS Senne Cornelis 1, Yannick Gansemans 1, Lieselot Deleye 1, Dieter Deforce 1,#,Filip Van Nieuwerburgh 1,#,* 1 Laboratory of Pharmaceutical
SOLiD Technology. library preparation & Sequencing Chemistry (sequencing by ligation!) Imaging and analysis. Application specific sample preparation
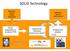 SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
SOLiD Technology Application specific sample preparation Application specific data analysis library preparation & emulsion PCR Sequencing Chemistry (sequencing by ligation!) Imaging and analysis SOLiD
Expression analysis of PIN genes in root tips and nodules of Lotus japonicus
 Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Article Expression analysis of PIN genes in root tips and nodules of Lotus japonicus Izabela Sańko-Sawczenko 1, Dominika Dmitruk 1, Barbara Łotocka 1, Elżbieta Różańska 1 and Weronika Czarnocka 1, * 1
Flowering time. Col C24 Cvi C24xCol C24xCvi ColxCvi
 Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Flowering time Rosette leaf number 50 45 40 35 30 25 20 15 10 5 0 Col C24 Cvi C24xCol C24xCvi ColxCvi Figure S1. Flowering time in three F 1 hybrids and their parental lines as measured by leaf number
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo
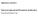 Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Supplementary materials to: Whole-mount single molecule FISH method for zebrafish embryo Yuma Oka and Thomas N. Sato Supplementary Figure S1. Whole-mount smfish with and without the methanol pretreatment.
Összefoglalás. Summary. Bevezetés
 A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
A talaj kálium ellátottságának vizsgálata módosított Baker-Amacher és,1 M CaCl egyensúlyi kivonószerek alkalmazásával Berényi Sándor Szabó Emese Kremper Rita Loch Jakab Debreceni Egyetem Agrár és Műszaki
7 th Iron Smelting Symposium 2010, Holland
 7 th Iron Smelting Symposium 2010, Holland Október 13-17 között került megrendezésre a Hollandiai Alphen aan den Rijn városában található Archeon Skanzenben a 7. Vasolvasztó Szimpózium. Az öt napos rendezvényen
7 th Iron Smelting Symposium 2010, Holland Október 13-17 között került megrendezésre a Hollandiai Alphen aan den Rijn városában található Archeon Skanzenben a 7. Vasolvasztó Szimpózium. Az öt napos rendezvényen
Új temékek az UD-GenoMed Kft. kínálatában!
 Új temékek az UD-GenMed Kft. kínálatában! Műanyag termékek: SARSTEDT műanyag termékek teljes választéka Egyszer használats labratóriumi műanyag eszközök szövet és sejttenyésztéshez Vérvételi és diagnsztikai
Új temékek az UD-GenMed Kft. kínálatában! Műanyag termékek: SARSTEDT műanyag termékek teljes választéka Egyszer használats labratóriumi műanyag eszközök szövet és sejttenyésztéshez Vérvételi és diagnsztikai
Suppl. Materials. Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids. Germany
 Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
Suppl. Materials Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids Stephanie Bresan 1, Anna Sznajder 1, Waldemar Hauf 2, Karl Forchhammer 2, Daniel Pfeiffer 3 and Dieter Jendrossek 1 1 Institute
The role of aristolochene synthase in diphosphate activation
 Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Supporting information for The role of aristolochene synthase in diphosphate activation Juan A. Faraldos, Verónica González and Rudolf K. Allemann * School of Chemistry, Cardiff University, Main Building,
Biológiai makromolekulák szerkezete
 Biológiai makromolekulák szerkezete Biomolekuláris nemkovalens kölcsönhatások Elektrosztatikus kölcsönhatások (sóhidak: 4-6 kcal/m, dipól-dipól: ~10-1 kcal/m Diszperziós erők (~10-2 kcal/m) Hidrogén hidak
Biológiai makromolekulák szerkezete Biomolekuláris nemkovalens kölcsönhatások Elektrosztatikus kölcsönhatások (sóhidak: 4-6 kcal/m, dipól-dipól: ~10-1 kcal/m Diszperziós erők (~10-2 kcal/m) Hidrogén hidak
A évi fizikai Nobel-díj
 A 2012. évi fizikai Nobel-díj "for ground-breaking experimental methods that enable measuring and manipulation of individual quantum systems" Serge Haroche David Wineland Ecole Normale Superieure, Párizs
A 2012. évi fizikai Nobel-díj "for ground-breaking experimental methods that enable measuring and manipulation of individual quantum systems" Serge Haroche David Wineland Ecole Normale Superieure, Párizs
KN-CP50. MANUAL (p. 2) Digital compass. ANLEITUNG (s. 4) Digitaler Kompass. GEBRUIKSAANWIJZING (p. 10) Digitaal kompas
 KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
KN-CP50 MANUAL (p. ) Digital compass ANLEITUNG (s. 4) Digitaler Kompass MODE D EMPLOI (p. 7) Boussole numérique GEBRUIKSAANWIJZING (p. 0) Digitaal kompas MANUALE (p. ) Bussola digitale MANUAL DE USO (p.
PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING PROPERTIES
 Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 371 379. PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING
Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 371 379. PLATTÍROZOTT ALUMÍNIUM LEMEZEK KÖTÉSI VISZONYAINAK TECHNOLÓGIAI VIZSGÁLATA TECHNOLOGICAL INVESTIGATION OF PLATED ALUMINIUM SHEETS BONDING
Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális
 Ipari Ökológia pp. 17 22. (2015) 3. évfolyam, 1. szám Magyar Ipari Ökológiai Társaság MIPOET 2015 Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális elegyekre* Tóth András
Ipari Ökológia pp. 17 22. (2015) 3. évfolyam, 1. szám Magyar Ipari Ökológiai Társaság MIPOET 2015 Extraktív heteroazeotróp desztilláció: ökologikus elválasztási eljárás nemideális elegyekre* Tóth András
Bagi Nyílászáró Gyártó és Kereskedelmi Kft.
 Bagi Nyílászáró Gyártó és Kereskedelmi Kft. 2191 BAG Jókai utca 13. Adószám: HU13390727-2-13 Cégjegyzék:13-09-101221 Telefon:+36-70-550-8452 Fax:+36-28-408-088 E-mail: bagifa@invitel.hu ksj.manufacturing@gmail.com
Bagi Nyílászáró Gyártó és Kereskedelmi Kft. 2191 BAG Jókai utca 13. Adószám: HU13390727-2-13 Cégjegyzék:13-09-101221 Telefon:+36-70-550-8452 Fax:+36-28-408-088 E-mail: bagifa@invitel.hu ksj.manufacturing@gmail.com
Búza László, M Schill Judit, Szentgyörgyi Mária, Ábrahám Ágnes, Debreczeni Lajos, Keresztúri József, Muránszky Géza. 2009. április 22.
 Gondolatok a MÉZ hamisítása kapcsán MIT-MIKOR-MELY Szereplő- MIKÉPPEN-MIÉRT hamisít, hamisíthat? Szofisztikált analitikai módszerek a hamisítások megelőzésére Búza László, M Schill Judit, Szentgyörgyi
Gondolatok a MÉZ hamisítása kapcsán MIT-MIKOR-MELY Szereplő- MIKÉPPEN-MIÉRT hamisít, hamisíthat? Szofisztikált analitikai módszerek a hamisítások megelőzésére Búza László, M Schill Judit, Szentgyörgyi
Cashback 2015 Deposit Promotion teljes szabályzat
 Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
Cashback 2015 Deposit Promotion teljes szabályzat 1. Definitions 1. Definíciók: a) Account Client s trading account or any other accounts and/or registers maintained for Számla Az ügyfél kereskedési számlája
NASODRILL ORRSPRAY: TARTÁLY- ÉS DOBOZFELIRAT, VALAMINT A BETEGTÁJÉKOZTATÓ SZÖVEGE. CSECSEMŐ GYERMEK FELNŐTT 100 ml-es üveg
 NASODRILL ORRSPRAY: TARTÁLY- ÉS DOBOZFELIRAT, VALAMINT A BETEGTÁJÉKOZTATÓ SZÖVEGE TARTÁLY - BOTTLE NASAL LAVAGE For chronic or recurring infection NASODRILL Formulated with thermal Luchon water naturally
NASODRILL ORRSPRAY: TARTÁLY- ÉS DOBOZFELIRAT, VALAMINT A BETEGTÁJÉKOZTATÓ SZÖVEGE TARTÁLY - BOTTLE NASAL LAVAGE For chronic or recurring infection NASODRILL Formulated with thermal Luchon water naturally
Szundikáló macska Sleeping kitty
 Model: Peter Budai 999. Diagrams: Peter Budai 999.. Oda-visszahajtás átlósan. Fold and unfold diagonally. 2. Behajtunk középre. Fold to the center. 3. Oda-visszahajtások derékszögben. Fold and unfold at
Model: Peter Budai 999. Diagrams: Peter Budai 999.. Oda-visszahajtás átlósan. Fold and unfold diagonally. 2. Behajtunk középre. Fold to the center. 3. Oda-visszahajtások derékszögben. Fold and unfold at
Mapping Sequencing Reads to a Reference Genome
 Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
Mapping Sequencing Reads to a Reference Genome High Throughput Sequencing RN Example applications: Sequencing a genome (DN) Sequencing a transcriptome and gene expression studies (RN) ChIP (chromatin immunoprecipitation)
PETER PAZMANY CATHOLIC UNIVERSITY Consortium members SEMMELWEIS UNIVERSITY, DIALOG CAMPUS PUBLISHER
 SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
SEMMELWEIS UNIVERSITY PETER PAMANY CATLIC UNIVERSITY Development of Complex Curricula for Molecular Bionics and Infobionics Programs within a consortial* framework** Consortium leader PETER PAMANY CATLIC
On The Number Of Slim Semimodular Lattices
 On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
On The Number Of Slim Semimodular Lattices Gábor Czédli, Tamás Dékány, László Ozsvárt, Nóra Szakács, Balázs Udvari Bolyai Institute, University of Szeged Conference on Universal Algebra and Lattice Theory
i1400 Image Processing Guide A-61623_zh-tw
 i1400 Image Processing Guide A-61623_zh-tw ................................................................. 1.............................................................. 1.........................................................
i1400 Image Processing Guide A-61623_zh-tw ................................................................. 1.............................................................. 1.........................................................
USER MANUAL Guest user
 USER MANUAL Guest user 1 Welcome in Kutatótér (Researchroom) Top menu 1. Click on it and the left side menu will pop up 2. With the slider you can make left side menu visible 3. Font side: enlarging font
USER MANUAL Guest user 1 Welcome in Kutatótér (Researchroom) Top menu 1. Click on it and the left side menu will pop up 2. With the slider you can make left side menu visible 3. Font side: enlarging font
LED UTCAI LÁMPATESTEK STREET LIGHTING
 LED UTCAI LÁMPATESTEK STREET LIGHTING LED Utcai lámpatestek Street Lights 1. Öntött ADC12 alumínium ház NOBEL porszórt festéssel 3. Edzett optikai lencsék irányított sugárzási szöggel 5. Meanwell meghajtó
LED UTCAI LÁMPATESTEK STREET LIGHTING LED Utcai lámpatestek Street Lights 1. Öntött ADC12 alumínium ház NOBEL porszórt festéssel 3. Edzett optikai lencsék irányított sugárzási szöggel 5. Meanwell meghajtó
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet Nonparametric Tests
 Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Nonparametric Tests Petra Petrovics Hypothesis Testing Parametric Tests Mean of a population Population proportion Population Standard Deviation Nonparametric Tests Test for Independence Analysis of Variance
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites
 Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
Markerless Escherichia coli rrn Deletion Strains for Genetic Determination of Ribosomal Binding Sites Selwyn Quan *, Ole Skovgaard, Robert E. McLaughlin, Ed T. Buurman,1, and Catherine L. Squires * Department
Téli kedvezményes ajánlataink
 Téli kedvezményes ajánlataink Ajánlataink 2017 január 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
Téli kedvezményes ajánlataink Ajánlataink 2017 január 31-ig érvényesek! BIOBASE műszerek a NucleotestBio Kft. kínálatában! Steril box-ok, lamináris box-ok: Hűtők, fagyasztók, ultramélyhűtők, autoklávok,
MELLÉKLET / ANNEX. EU MEGFELELŐSÉGI NYILATKOZAT-hoz for EU DECLARATION OF CONFORMITY
 TRACON BUDAPEST KFT. TRACON BUDAPEST LTD. 2120 DUNAKESZI, PALLAG U. 23. TEL.: (27) 540-000, FAX: (27) 540-005 WWW.TRACON.HU EU MEGFELELŐSÉGI NYILATKOZAT, a 79/1997. (XII. 31.) IKIM számú és a 374/2012.
TRACON BUDAPEST KFT. TRACON BUDAPEST LTD. 2120 DUNAKESZI, PALLAG U. 23. TEL.: (27) 540-000, FAX: (27) 540-005 WWW.TRACON.HU EU MEGFELELŐSÉGI NYILATKOZAT, a 79/1997. (XII. 31.) IKIM számú és a 374/2012.
Laborvegyszer árlista
 Laborvegyszer árlista Érvényesség: 2018.03.01.-től visszavonásig H-1131 Budapest, Madarász V. u. 1/A. Központi iroda/postacím: 1141 Budapest, Fogarasi út 218. Telefon: +36-1-273-13-78 Fax: +36-1-273-13-79
Laborvegyszer árlista Érvényesség: 2018.03.01.-től visszavonásig H-1131 Budapest, Madarász V. u. 1/A. Központi iroda/postacím: 1141 Budapest, Fogarasi út 218. Telefon: +36-1-273-13-78 Fax: +36-1-273-13-79
Pro sensors Measurement sensors to IP Thermo Professional network
 Pro sensors Measurement sensors to IP Thermo Professional network T-05 Temperature sensor TH-05 Temperature, humidity sensor THP- 05 Temperature, humidity, air pressure, air velocity, wet sensors indoor
Pro sensors Measurement sensors to IP Thermo Professional network T-05 Temperature sensor TH-05 Temperature, humidity sensor THP- 05 Temperature, humidity, air pressure, air velocity, wet sensors indoor
Vitorláshal Angelfish
 Model: Peter udai 996. Diagrams: Peter udai 998.. Oda-visszahajtás felezve. Fordítsd meg a papírt! Fold in half and unfold. Turn the paper over. 2. Oda-visszahajtás átlósan. Fold and unfold diagonally.
Model: Peter udai 996. Diagrams: Peter udai 998.. Oda-visszahajtás felezve. Fordítsd meg a papírt! Fold in half and unfold. Turn the paper over. 2. Oda-visszahajtás átlósan. Fold and unfold diagonally.
UFS Productspecification
 Page 1 of 5 General Information Dry mix for 'Potato Flakes with Milk' - the mixture contains skimmed milk powder - solely water is necessary for the preparation of the potato mash - bulk density 380-420
Page 1 of 5 General Information Dry mix for 'Potato Flakes with Milk' - the mixture contains skimmed milk powder - solely water is necessary for the preparation of the potato mash - bulk density 380-420
Kezdőlap > Termékek > Szabályozó rendszerek > EASYLAB és TCU-LON-II szabályozó rendszer LABCONTROL > Érzékelő rendszerek > Típus DS-TRD-01
 Típus DS-TRD FOR EASYLAB FUME CUPBOARD CONTROLLERS Sash distance sensor for the variable, demand-based control of extract air flows in fume cupboards Sash distance measurement For fume cupboards with vertical
Típus DS-TRD FOR EASYLAB FUME CUPBOARD CONTROLLERS Sash distance sensor for the variable, demand-based control of extract air flows in fume cupboards Sash distance measurement For fume cupboards with vertical
Széchenyi István Egyetem www.sze.hu/~herno
 Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
Oldal: 1/6 A feladat során megismerkedünk a C# és a LabVIEW összekapcsolásának egy lehetőségével, pontosabban nagyon egyszerű C#- ban írt kódból fordítunk DLL-t, amit meghívunk LabVIEW-ból. Az eljárás
Good-One Mfg., LLC. We didn t invent barbeque We just perfected it. [Type text] Performance Testing of Rodeo Model Good-One Smoker August 29, 2008
![Good-One Mfg., LLC. We didn t invent barbeque We just perfected it. [Type text] Performance Testing of Rodeo Model Good-One Smoker August 29, 2008 Good-One Mfg., LLC. We didn t invent barbeque We just perfected it. [Type text] Performance Testing of Rodeo Model Good-One Smoker August 29, 2008](/thumbs/96/126904219.jpg) [Type text] Good-One Mfg., LLC Performance Testing of Rodeo Model Good-One Smoker August 29, 2008 We didn t invent barbeque We just perfected it Chris Marks 9-Time BBQ World Champion Table of Contents
[Type text] Good-One Mfg., LLC Performance Testing of Rodeo Model Good-One Smoker August 29, 2008 We didn t invent barbeque We just perfected it Chris Marks 9-Time BBQ World Champion Table of Contents
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon
 Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Hibridspecifikus tápanyag-és vízhasznosítás kukoricánál csernozjom talajon Karancsi Lajos Gábor Debreceni Egyetem Agrár és Gazdálkodástudományok Centruma Mezőgazdaság-, Élelmiszertudományi és Környezetgazdálkodási
Cluster Analysis. Potyó László
 Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Cluster Analysis Potyó László What is Cluster Analysis? Cluster: a collection of data objects Similar to one another within the same cluster Dissimilar to the objects in other clusters Cluster analysis
Supplemental Information. RNase H1-Dependent Antisense Oligonucleotides. Are Robustly Active in Directing RNA Cleavage
 YMTHE, Volume 25 Supplemental Information RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus Xue-Hai Liang, Hong Sun, Joshua
YMTHE, Volume 25 Supplemental Information RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus Xue-Hai Liang, Hong Sun, Joshua
Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley M. Lemon, Lawrence M. Pfeffer, Kui Li
 Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Supplemental Material IRF3-dependent and NF- B-independent viral induction of the zinc-finger antiviral protein Nan Wang, Qingming Dong, Jingjing Li, Rohit K. Jangra, Meiyun Fan, Allan R. Brasier, Stanley
Supporting Information
 Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Supporting Information Plasmid Construction DNA cloning was carried out according to standard protocols. The sequence of all plasmids was confirmed by DNA sequencing and amplified using the E. coli strain
Résbefúvó anemosztátok méréses vizsgálata érintõleges légvezetési rendszer alkalmazása esetén
 Résbefúvó anemostátok méréses visgálata érintõleges légveetési rendser alkalmaása esetén Both Balás 1 Goda Róbert 2 Abstract The use of slot diffusers in tangential air supply systems is widespread not
Résbefúvó anemostátok méréses visgálata érintõleges légveetési rendser alkalmaása esetén Both Balás 1 Goda Róbert 2 Abstract The use of slot diffusers in tangential air supply systems is widespread not
TDA-TAR ÉS O-TDA FOLYADÉKÁRAMOK ELEGYÍTHETŐSÉGÉNEK VIZSGÁLATA STUDY OF THE MIXABILITY OF TDA-TAR AND O-TDA LIQUID STREAMS
 Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 147 156. TDA-TAR ÉS O-TDA FOLYADÉKÁRAMOK ELEGYÍTHETŐSÉGÉNEK VIZSGÁLATA STUDY OF THE MIXABILITY OF TDA-TAR AND O-TDA LIQUID STREAMS HUTKAINÉ GÖNDÖR
Anyagmérnöki Tudományok, 37. kötet, 1. szám (2012), pp. 147 156. TDA-TAR ÉS O-TDA FOLYADÉKÁRAMOK ELEGYÍTHETŐSÉGÉNEK VIZSGÁLATA STUDY OF THE MIXABILITY OF TDA-TAR AND O-TDA LIQUID STREAMS HUTKAINÉ GÖNDÖR
Miskolci Egyetem Gazdaságtudományi Kar Üzleti Információgazdálkodási és Módszertani Intézet. Nonparametric Tests. Petra Petrovics.
 Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
Nonparametric Tests Petra Petrovics PhD Student Hypothesis Testing Parametric Tests Mean o a population Population proportion Population Standard Deviation Nonparametric Tests Test or Independence Analysis
TÁMOPͲ4.2.2.AͲ11/1/KONVͲ2012Ͳ0029
 AUTOTECH Jármipari anyagfejlesztések: célzott alapkutatás az alakíthatóság, hkezelés és hegeszthetség témaköreiben TÁMOP4.2.2.A11/1/KONV20120029 www.autotech.unimiskolc.hu ANYAGSZERKEZETTANI ÉS ANYAGTECHNOLÓGIAI
AUTOTECH Jármipari anyagfejlesztések: célzott alapkutatás az alakíthatóság, hkezelés és hegeszthetség témaköreiben TÁMOP4.2.2.A11/1/KONV20120029 www.autotech.unimiskolc.hu ANYAGSZERKEZETTANI ÉS ANYAGTECHNOLÓGIAI
9el[hW][e\L;BI IjWdZWhZi
![9el[hW][e\L;BI IjWdZWhZi 9el[hW][e\L;BI IjWdZWhZi](/thumbs/102/156771080.jpg) 9el[hW][e\L;BI IjWdZWhZi The content and activities in Alive 3 and Alive 4 have been prepared to allow students to achieve the Victorian Essential Learning Standards (VELS) for Level 6. The key elements
9el[hW][e\L;BI IjWdZWhZi The content and activities in Alive 3 and Alive 4 have been prepared to allow students to achieve the Victorian Essential Learning Standards (VELS) for Level 6. The key elements
DECLARATION OF PERFORMANCE No. GST REV 1.03 According to Construction Products Regulation EU No. 305/2011
 DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
DECLARATION OF PERFORMANCE No. According to Construction Products Regulation EU No. 305/2011 This declaration is available in the following languages: English Declaration of Performance Page 2-3 Hungarian
építészet & design ipari alkalmazás teherautó felépítmény
 A Design-Composit egy kompozitpaneleket gyártó vállalat, mely teherautó felépítményekhez, az építészet számára és design termékekhez készít paneleket. We are an innovative manufacturer of composite panels
A Design-Composit egy kompozitpaneleket gyártó vállalat, mely teherautó felépítményekhez, az építészet számára és design termékekhez készít paneleket. We are an innovative manufacturer of composite panels
Model Identification and Predictive Control of a Laboratory Binary Distillation Column
 Model Identification and Predictive Control of a Laboratory Binary Distillation Column Ján Drgoňa, Martin Klaučo, Richard Valo, Jakub Bendžala, and Miroslav Fikar Slovak University of Technology in Bratislava,
Model Identification and Predictive Control of a Laboratory Binary Distillation Column Ján Drgoňa, Martin Klaučo, Richard Valo, Jakub Bendžala, and Miroslav Fikar Slovak University of Technology in Bratislava,
Phenotype. Genotype. It is like any other experiment! What is a bioinformatics experiment? Remember the Goal. Infectious Disease Paradigm
 It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
It is like any other experiment! What is a bioinformatics experiment? You need to know your data/input sources You need to understand your methods and their assumptions You need a plan to get from point
4 vana, vanb, vanc1, vanc2
 77 1) 2) 2) 3) 4) 5) 1) 2) 3) 4) 5) 16 12 7 17 4 8 2003 1 2004 7 3 Enterococcus 5 Enterococcus faecalis 1 Enterococcus avium 1 Enterococcus faecium 2 5 PCR E. faecium 1 E-test MIC 256 mg/ml vana MRSA MRSA
77 1) 2) 2) 3) 4) 5) 1) 2) 3) 4) 5) 16 12 7 17 4 8 2003 1 2004 7 3 Enterococcus 5 Enterococcus faecalis 1 Enterococcus avium 1 Enterococcus faecium 2 5 PCR E. faecium 1 E-test MIC 256 mg/ml vana MRSA MRSA
A modern e-learning lehetőségei a tűzoltók oktatásának fejlesztésében. Dicse Jenő üzletfejlesztési igazgató
 A modern e-learning lehetőségei a tűzoltók oktatásának fejlesztésében Dicse Jenő üzletfejlesztési igazgató How to apply modern e-learning to improve the training of firefighters Jenő Dicse Director of
A modern e-learning lehetőségei a tűzoltók oktatásának fejlesztésében Dicse Jenő üzletfejlesztési igazgató How to apply modern e-learning to improve the training of firefighters Jenő Dicse Director of
Using the CW-Net in a user defined IP network
 Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
Using the CW-Net in a user defined IP network Data transmission and device control through IP platform CW-Net Basically, CableWorld's CW-Net operates in the 10.123.13.xxx IP address range. User Defined
mintasepcifikus mikrokapilláris elektroforézis Lab-on-Chip elektroforézis / elektrokinetikus elven DNS, RNS, mirns 12, fehérje 10, sejtes minta 6
 Agilent 2100 Bioanalyzer mikrokapilláris gélelektroforézis rendszer G2943CA 2100 Bioanalyzer system forgalmazó: Kromat Kft. 1112 Budapest Péterhegyi u. 98. t:36 (1) 248-2110 www.kromat.hu bio@kromat.hu
Agilent 2100 Bioanalyzer mikrokapilláris gélelektroforézis rendszer G2943CA 2100 Bioanalyzer system forgalmazó: Kromat Kft. 1112 Budapest Péterhegyi u. 98. t:36 (1) 248-2110 www.kromat.hu bio@kromat.hu
DECLARATION OF PERFORMANCE. No. CPR-20-IC-204
 Page 1 of 3 DECLARATION OF PERFORMANCE No. CPR-20-IC-204 1. Unique identification code of the product-type: 1000003- Uponor Tacker Panel Roll 30-3 1000009- Uponor Tacker Panel 30-3 1000004- Uponor Tacker
Page 1 of 3 DECLARATION OF PERFORMANCE No. CPR-20-IC-204 1. Unique identification code of the product-type: 1000003- Uponor Tacker Panel Roll 30-3 1000009- Uponor Tacker Panel 30-3 1000004- Uponor Tacker
AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN
 Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
Tájökológiai Lapok 5 (2): 287 293. (2007) 287 AZ ERDÕ NÖVEKEDÉSÉNEK VIZSGÁLATA TÉRINFORMATIKAI ÉS FOTOGRAMMETRIAI MÓDSZEREKKEL KARSZTOS MINTATERÜLETEN ZBORAY Zoltán Honvédelmi Minisztérium Térképészeti
Elektrofiziológiai vizsgálómódszerek alkalmazása a sejtek elektromos tevékenységének kutatásában. A kezdetek 1.
 Elektrofiziológiai vizsgálómódszerek alkalmazása a sejtek elektromos tevékenységének kutatásában A kezdetek 1. 1935-1952 A klasszikus membrán biofizika megteremtése membránpermeabilitás kinetikája, membránfehérjék
Elektrofiziológiai vizsgálómódszerek alkalmazása a sejtek elektromos tevékenységének kutatásában A kezdetek 1. 1935-1952 A klasszikus membrán biofizika megteremtése membránpermeabilitás kinetikája, membránfehérjék
Supplemental Information. Investigation of Penicillin Binding Protein. (PBP)-like Peptide Cyclase and Hydrolase
 Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
Cell Chemical Biology, Volume upplemental Information Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and ydrolase in urugamide on-ribosomal Peptide Biosynthesis Yongjun Zhou, Xiao
Computer Architecture
 Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
Computer Architecture Locality-aware programming 2016. április 27. Budapest Gábor Horváth associate professor BUTE Department of Telecommunications ghorvath@hit.bme.hu Számítógép Architektúrák Horváth
AZ ÓLOM MOBILIZÁLHATÓSÁGÁNAK VIZSGÁLATA ÓLOMÜVEGEKBŐL DETERMINATION OF MOBILISATION OF LEAD FROM HIGH LEAD CONTENT GLASSES
 Anyagmérnöki Tudományok, 38/1. (2013), pp. 165 172. AZ ÓLOM MOBILIZÁLHATÓSÁGÁNAK VIZSGÁLATA ÓLOMÜVEGEKBŐL DETERMINATION OF MOBILISATION OF LEAD FROM HIGH LEAD CONTENT GLASSES LAKATOS JÁNOS 1 MUCSI GÁBOR
Anyagmérnöki Tudományok, 38/1. (2013), pp. 165 172. AZ ÓLOM MOBILIZÁLHATÓSÁGÁNAK VIZSGÁLATA ÓLOMÜVEGEKBŐL DETERMINATION OF MOBILISATION OF LEAD FROM HIGH LEAD CONTENT GLASSES LAKATOS JÁNOS 1 MUCSI GÁBOR
TIOLKARBAMÁT TÍPUSÚ NÖVÉNYVÉDŐ SZER HATÓANYAGOK ÉS SZÁRMAZÉKAIK KÉMIAI OXIDÁLHATÓSÁGÁNAK VIZSGÁLATA I
 Műszaki Földtudományi Közlemények, 83. kötet, 1. szám (2012), pp. 137 146. TIOLKARBAMÁT TÍPUSÚ NÖVÉNYVÉDŐ SZER HATÓANYAGOK ÉS SZÁRMAZÉKAIK KÉMIAI OXIDÁLHATÓSÁGÁNAK VIZSGÁLATA I. S-ETIL-N,N-DI-N-PROPIL-TIOLKARBAMÁT
Műszaki Földtudományi Közlemények, 83. kötet, 1. szám (2012), pp. 137 146. TIOLKARBAMÁT TÍPUSÚ NÖVÉNYVÉDŐ SZER HATÓANYAGOK ÉS SZÁRMAZÉKAIK KÉMIAI OXIDÁLHATÓSÁGÁNAK VIZSGÁLATA I. S-ETIL-N,N-DI-N-PROPIL-TIOLKARBAMÁT
Dynamic freefly DIVE POOL LINES
 Dynamic freefly 3.rész Part 3 DIVE POOL LINES A dynamic repülés három fajta mozgásból áll: Dynamic freeflying is composed of three types of movements: -Lines (vízszintes "szlalom") (horizontal "slalom")
Dynamic freefly 3.rész Part 3 DIVE POOL LINES A dynamic repülés három fajta mozgásból áll: Dynamic freeflying is composed of three types of movements: -Lines (vízszintes "szlalom") (horizontal "slalom")
Implementation of water quality monitoring
 Joint Ipoly/Ipel Catchment Management HUSK/1101/2.1.1/0153 Implementation of water quality monitoring Dr. Adrienne Clement clement@vkkt.bme.hu Budapest University of Technology and Economics Department
Joint Ipoly/Ipel Catchment Management HUSK/1101/2.1.1/0153 Implementation of water quality monitoring Dr. Adrienne Clement clement@vkkt.bme.hu Budapest University of Technology and Economics Department
Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali. Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang*
 Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Supplementary Information Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali Zhiyuan Yin, Baitao Zhu, Hao Feng, Lili Huang* State Key Laboratory of Crop
Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter
 Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter DOI: http://doi.org/10.13140/rg.2.2.28994.22721 A tudományos közlemények írása minden szakma művelésének
Néhány folyóiratkereső rendszer felsorolása és példa segítségével vázlatos bemutatása Sasvári Péter DOI: http://doi.org/10.13140/rg.2.2.28994.22721 A tudományos közlemények írása minden szakma művelésének
«DURAK HAZELNUTS» PRESENTATION
 «DURAK HAZELNUTS» PRESENTATION PRODUCT RANGE CAPACITIES p.a. HAZELNUT KERNELS RAW : 80.000 MT ------------------------------------------------- ROASTINGCAPACITY : 30.000 MT, out of which: ROASTED WHOLE
«DURAK HAZELNUTS» PRESENTATION PRODUCT RANGE CAPACITIES p.a. HAZELNUT KERNELS RAW : 80.000 MT ------------------------------------------------- ROASTINGCAPACITY : 30.000 MT, out of which: ROASTED WHOLE
General information for the participants of the GTG Budapest, 2017 meeting
 General information for the participants of the GTG Budapest, 2017 meeting Currency is Hungarian Forint (HUF). 1 EUR 310 HUF, 1000 HUF 3.20 EUR. Climate is continental, which means cold and dry in February
General information for the participants of the GTG Budapest, 2017 meeting Currency is Hungarian Forint (HUF). 1 EUR 310 HUF, 1000 HUF 3.20 EUR. Climate is continental, which means cold and dry in February
Lexington Public Schools 146 Maple Street Lexington, Massachusetts 02420
 146 Maple Street Lexington, Massachusetts 02420 Surplus Printing Equipment For Sale Key Dates/Times: Item Date Time Location Release of Bid 10/23/2014 11:00 a.m. http://lps.lexingtonma.org (under Quick
146 Maple Street Lexington, Massachusetts 02420 Surplus Printing Equipment For Sale Key Dates/Times: Item Date Time Location Release of Bid 10/23/2014 11:00 a.m. http://lps.lexingtonma.org (under Quick
For the environmentally aware
 Környezet, tudatos embereknek or the environmentally aware A k3 a Könyves Kálmán körúton, a főváros egyik legdinamikusabban fejlődő területén felépülő új A kategóriás fenntartható ház, amelynek legfőbb
Környezet, tudatos embereknek or the environmentally aware A k3 a Könyves Kálmán körúton, a főváros egyik legdinamikusabban fejlődő területén felépülő új A kategóriás fenntartható ház, amelynek legfőbb
A golyók felállítása a Pool-biliárd 8-as játékának felel meg. A golyók átmérıje 57.2 mm. 15 számozott és egy fehér golyó. Az elsı 7 egyszínő, 9-15-ig
 A golyók elhelyezkedése a Snooker alaphelyzetet mutatja. A golyók átmérıje 52 mm, egyszínőek. 15 db piros, és 1-1 db fehér, fekete, rózsa, kék, barna, zöld, sárga. A garázsban állítjuk fel, ilyenkor az
A golyók elhelyezkedése a Snooker alaphelyzetet mutatja. A golyók átmérıje 52 mm, egyszínőek. 15 db piros, és 1-1 db fehér, fekete, rózsa, kék, barna, zöld, sárga. A garázsban állítjuk fel, ilyenkor az
MINO V2 ÁLLVÁNY CSERÉJE V4-RE
 MINO V2 remote controlled MINO V2 ÁLLVÁNY CSERÉJE V4-RE Mino V3 circuit board replacement Mino V2-V4 csere készlet ezüst Art# 59348S, Mino V2-V4 csere készlet fehér Art# 59348W V4 áramköri lap Art# 75914
MINO V2 remote controlled MINO V2 ÁLLVÁNY CSERÉJE V4-RE Mino V3 circuit board replacement Mino V2-V4 csere készlet ezüst Art# 59348S, Mino V2-V4 csere készlet fehér Art# 59348W V4 áramköri lap Art# 75914
Márkaépítés a YouTube-on
 Márkaépítés a YouTube-on Tv+ Adj hozzá YouTube-ot, Google Ground, 2016 Március 7. Bíró Pál, Google - YouTube 9,000,000 INTERNETTEL BÍRÓ ESZKÖZÖK VOLUMENE GLOBÁLISAN WEARABLES OKOS TV 8,000,000 7,000,000
Márkaépítés a YouTube-on Tv+ Adj hozzá YouTube-ot, Google Ground, 2016 Március 7. Bíró Pál, Google - YouTube 9,000,000 INTERNETTEL BÍRÓ ESZKÖZÖK VOLUMENE GLOBÁLISAN WEARABLES OKOS TV 8,000,000 7,000,000
SZÉKEK & FOTELEK CHAIRS & ARMCHAIRS
 SZÉKEK & FOTELEK CHAIRS & ARMCHAIRS 1 Art&Deco Contemporary is a retail and interior design enterprise in one. As for our interior design department, we are focused on furniture design, and Art&Deco s
SZÉKEK & FOTELEK CHAIRS & ARMCHAIRS 1 Art&Deco Contemporary is a retail and interior design enterprise in one. As for our interior design department, we are focused on furniture design, and Art&Deco s
Csatlakozás a BME eduroam hálózatához Setting up the BUTE eduroam network
 Csatlakozás a BME eduroam hálózatához Setting up the BUTE eduroam network Table of Contents Windows 7... 2 Windows 8... 6 Windows Phone... 11 Android... 12 iphone... 14 Linux (Debian)... 20 Sebők Márton
Csatlakozás a BME eduroam hálózatához Setting up the BUTE eduroam network Table of Contents Windows 7... 2 Windows 8... 6 Windows Phone... 11 Android... 12 iphone... 14 Linux (Debian)... 20 Sebők Márton
Utasítások. Üzembe helyezés
 HASZNÁLATI ÚTMUTATÓ Üzembe helyezés Utasítások Windows XP / Vista / Windows 7 / Windows 8 rendszerben történő telepítéshez 1 Töltse le az AORUS makróalkalmazás telepítőjét az AORUS hivatalos webhelyéről.
HASZNÁLATI ÚTMUTATÓ Üzembe helyezés Utasítások Windows XP / Vista / Windows 7 / Windows 8 rendszerben történő telepítéshez 1 Töltse le az AORUS makróalkalmazás telepítőjét az AORUS hivatalos webhelyéről.
± ± ± ƒ ± ± ± ± ± ± ± ƒ. ± ± ƒ ± ± ± ± ƒ. ± ± ± ± ƒ
 ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ç å ± ƒ ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ä ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ±
± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ç å ± ƒ ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ä ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ± ƒ ± ± ± ± ± ƒ ± ± ± ± ƒ ± ± ± ƒ ± ± ƒ ± ± ± ± ± ± ± ± ± ± ± ± ± ±
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel
 Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
Angol Középfokú Nyelvvizsgázók Bibliája: Nyelvtani összefoglalás, 30 kidolgozott szóbeli tétel, esszé és minta levelek + rendhagyó igék jelentéssel Timea Farkas Click here if your download doesn"t start
Zephyr használati utasítás
 Zephyr használati utasítás fontos óvintézkedések Ez a termék háztartási használatra készült. Elektromos termékek használatánál, különösen gyermekek jelenlétében, mindig be kell tartani a következő alapvető
Zephyr használati utasítás fontos óvintézkedések Ez a termék háztartási használatra készült. Elektromos termékek használatánál, különösen gyermekek jelenlétében, mindig be kell tartani a következő alapvető
Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel
 Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel /Study of Solvent Gradient by Simulated Moving Bed Preparative Liqiud Chpomatography Technology/ 1 Nagy
Oldószer Gradiensek Vizsgálata Szimulált Mozgóréteges Preparatív Folyafékkromatográfiás Művelettel /Study of Solvent Gradient by Simulated Moving Bed Preparative Liqiud Chpomatography Technology/ 1 Nagy
Közlekedésbiztonsági Szakmai Nap
 Közlekedésbiztonsági Szakmai Nap 2011. November 23. 1 Svinger László üzletágvezető 3M. Minnesota Mining & Manufacturing 2 3M technológiák Ab Abrasives Ac Acoustics Ad Adhesives Am Advanced Materials An
Közlekedésbiztonsági Szakmai Nap 2011. November 23. 1 Svinger László üzletágvezető 3M. Minnesota Mining & Manufacturing 2 3M technológiák Ab Abrasives Ac Acoustics Ad Adhesives Am Advanced Materials An
PRECÍZIÓS CSÁVÁZÓGÉP VETŐMAG FELDOLGOZÁS SZÁLLÍTÓ ESZKÖZÖK SZÁRÍTÓK VETŐMAG FELDOLGOZÁS ELEKTRONIKUS OSZTÁLYOZÁS TÁROLÁS KULCSRAKÉSZ ÜZEMEK
 HU PRECÍZIÓS CSÁVÁZÓGÉP VETŐMAG FELDOLGOZÁS SZÁLLÍTÓ ESZKÖZÖK SZÁRÍTÓK VETŐMAG FELDOLGOZÁS ELEKTRONIKUS OSZTÁLYOZÁS TÁROLÁS KULCSRAKÉSZ ÜZEMEK HATÉKONY CSÁVÁZÓANYAG FELHASZNÁLÁS MODERN CSÁVÁZÁSI TECHNOLÓGIA
HU PRECÍZIÓS CSÁVÁZÓGÉP VETŐMAG FELDOLGOZÁS SZÁLLÍTÓ ESZKÖZÖK SZÁRÍTÓK VETŐMAG FELDOLGOZÁS ELEKTRONIKUS OSZTÁLYOZÁS TÁROLÁS KULCSRAKÉSZ ÜZEMEK HATÉKONY CSÁVÁZÓANYAG FELHASZNÁLÁS MODERN CSÁVÁZÁSI TECHNOLÓGIA
Formula Sound árlista
 MIXERS FF-6000; FF6000P Formula Sound 160 6 channel dual format DJ mixer with removable fader panel. (Supplied with linear faders) Formula Sound 160P As above but with PRO X crossfade fitted. Formula Sound
MIXERS FF-6000; FF6000P Formula Sound 160 6 channel dual format DJ mixer with removable fader panel. (Supplied with linear faders) Formula Sound 160P As above but with PRO X crossfade fitted. Formula Sound
Tudományos Ismeretterjesztő Társulat
 Sample letter number 3. Russell Ltd. 57b Great Hawthorne Industrial Estate Hull East Yorkshire HU 19 5BV 14 Bebek u. Budapest H-1105 10 December, 2009 Ref.: complaint Dear Sir/Madam, After seeing your
Sample letter number 3. Russell Ltd. 57b Great Hawthorne Industrial Estate Hull East Yorkshire HU 19 5BV 14 Bebek u. Budapest H-1105 10 December, 2009 Ref.: complaint Dear Sir/Madam, After seeing your
The Measurement of The Three Components of The Cutting Force During The Turning Process
 The Measurement of The Three Components of The Cutting Force During The Turning Process CS. NEMES 1, L. JAVOREK 2, S. BODZÁS 3, S. PÁLINKÁS 4 1 Department of Mechanical Engineering, Faculty of Engineering,
The Measurement of The Three Components of The Cutting Force During The Turning Process CS. NEMES 1, L. JAVOREK 2, S. BODZÁS 3, S. PÁLINKÁS 4 1 Department of Mechanical Engineering, Faculty of Engineering,
ANNEX V / V, MELLÉKLET
 ANNEX V / V, MELLÉKLET VETERINARY CERTIFICATE EMBRYOS OF DOMESTIC ANIMALS OF THE BOVINE SPECIES FOR IMPORTS COLLECTED OR PRODUCED BEFORE 1 st JANUARY 2006 ÁLLAT-EGÉSZSÉGÜGYI BIZONYÍTVÁNY 2006. JANUÁR 1-JE
ANNEX V / V, MELLÉKLET VETERINARY CERTIFICATE EMBRYOS OF DOMESTIC ANIMALS OF THE BOVINE SPECIES FOR IMPORTS COLLECTED OR PRODUCED BEFORE 1 st JANUARY 2006 ÁLLAT-EGÉSZSÉGÜGYI BIZONYÍTVÁNY 2006. JANUÁR 1-JE
TRENDnetVIEW Pro szoftvert. ŸGyors telepítési útmutató (1)
 TRENDnetVIEW Pro szoftvert ŸGyors telepítési útmutató (1) TRENDnetVIEW Pro/05.29.2014 Tartalomjegyzék TRENDnetVIEW Pro Management Software követelmények... 13 TRENDnetVIEW Pro Telepítése... 14 Videokamerák
TRENDnetVIEW Pro szoftvert ŸGyors telepítési útmutató (1) TRENDnetVIEW Pro/05.29.2014 Tartalomjegyzék TRENDnetVIEW Pro Management Software követelmények... 13 TRENDnetVIEW Pro Telepítése... 14 Videokamerák
Geokémia gyakorlat. 1. Geokémiai adatok értelmezése: egyszerű statisztikai módszerek. Geológus szakirány (BSc) Dr. Lukács Réka
 Geokémia gyakorlat 1. Geokémiai adatok értelmezése: egyszerű statisztikai módszerek Geológus szakirány (BSc) Dr. Lukács Réka MTA-ELTE Vulkanológiai Kutatócsoport e-mail: reka.harangi@gmail.com ALAPFOGALMAK:
Geokémia gyakorlat 1. Geokémiai adatok értelmezése: egyszerű statisztikai módszerek Geológus szakirány (BSc) Dr. Lukács Réka MTA-ELTE Vulkanológiai Kutatócsoport e-mail: reka.harangi@gmail.com ALAPFOGALMAK:
